In Part 1 of this series we covered some of the basic functions that we will need to perform resampling and simulations in R.
In Part 2 we will now move to building bootstrap resamples and simulating distributions for both bivariate and multivariate relationships. Some stuff we will cover:
- Coding bootstrap resampling by hand for both a single variable (mean) and regression coefficients.
- Using the boot() function from the boot package to perform bootstrapping for both a single variable and regression coefficients.
- Creating a simulation where two variables are in some way dependent on each other (correlate).
- Creating a simulation where multiple variables are correlated with each other.
- Finally, to understand what is going on with these simulated distributions we will also work through code that shows us the relationship between variables using both covariance and correlation matrices.
As always, all code is freely available in the Github repository.
Resampling
The resampling approach we will use here is bootstrapping. The general concept of bootstrapping is as follows:
- Draw multiple random samples from observed data with replacement.
- Draws must be independent and each observation must have an equal chance of being selected.
- The bootstrap sample should be the same size as the observed data in order to use as much information from the sample as possible.
- Calculate the mean resampled data and store it.
- Repeat this process thousands of times and summarize the mean of resampled means and the standard deviation of resampled means to obtain summary statistics of your bootstrapped resamples.
Write the bootstrap resampling by hand
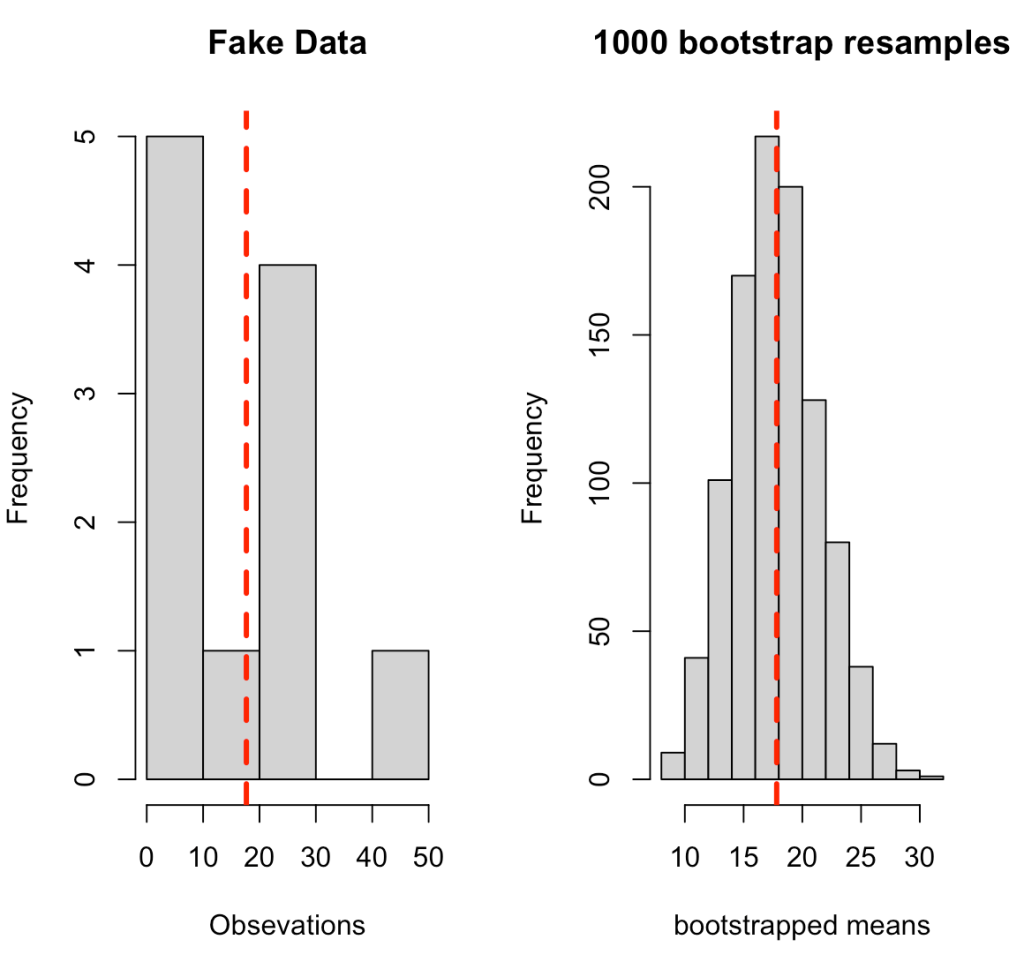
The code below goes through the process of creating some fake data and then writing a for() loop that produces 1000 bootstrap resamples. The for() loop was introduced in Part 1 of this series. In a nutshell, we are taking a random sample from the fake data (with replacement), calculating the mean of that random sample, and storing it in the element boot_dat. From there, we calculate the summary statistics or the original sample and the bootstrap resample, which we store in a data frame for comparison purposes, and then produce a histogram of the original sample and the bootstrap resample, which are visualized below the code.
library(tidyverse)
## create fake data
dat <- c(5, 10, 44, 3, 29, 10, 16.7, 22.3, 28, 1.4, 25)
### Bootstrap Resamples ###
# we want 1000 bootstrap resamples
n_boots <- 1000
## create an empty vector to store our bootstraps
boot_dat <- rep(NA, n_boots)
# set seed for reproducibility
set.seed(786)
# write for() loop for the resampling
for(i in 1:n_boots){
# random sample of 1:n number of observations in our data, with replacement
ind <- sample(1:length(dat), replace = TRUE)
# Use the row indexes to select the given values from the vector and calculate the mean
boot_dat[i] <- mean(dat[ind])
}
# Look at the first 6 bootstrapped means
head(boot_dat)
### Compare Bootstrap data to original data ###
## mean and standard deviation of the fake data
dat_mean <- mean(dat)
dat_sd <- sd(dat)
# standard error of the mean
dat_se <- sd(dat) / sqrt(length(dat))
# 95% confidence interval
dat_ci95 <- paste0(round(dat_mean - 1.96*dat_se, 1), ", ", round(dat_mean + 1.96*dat_se, 1))
# mean an SD of bootstrapped data
boot_mean <- mean(boot_dat)
# the vector is the mean of each bootstrap sample, so the standard deviation of these means represents the standard error
boot_se <- sd(boot_dat)
# to get the standard deviation we can convert the standard error back
boot_sd <- boot_se * sqrt(length(dat))
# 95% quantile interval
boot_ci95 <- paste0(round(boot_mean - 1.96*boot_se, 1), ", ", round(boot_mean + 1.96*boot_se, 1)) ## Put everything together data.frame(data = c("fake sample", "bootstrapped resamples"), N = c(length(dat), length(boot_dat)), mean = c(dat_mean, boot_mean), sd = c(dat_sd, boot_sd), se = c(dat_se, boot_se), ci95 = c(dat_ci95, boot_ci95)) %>%
knitr::kable()
# plot the distributions
par(mfrow = c(1, 2))
hist(dat,
xlab = "Obsevations",
main = "Fake Data")
abline(v = dat_mean,
col = "red",
lwd = 3,
lty = 2)
hist(boot_dat,
xlab = "bootstrapped means",
main = "1000 bootstrap resamples")
abline(v = boot_mean,
col = "red",
lwd = 3,
lty = 2)
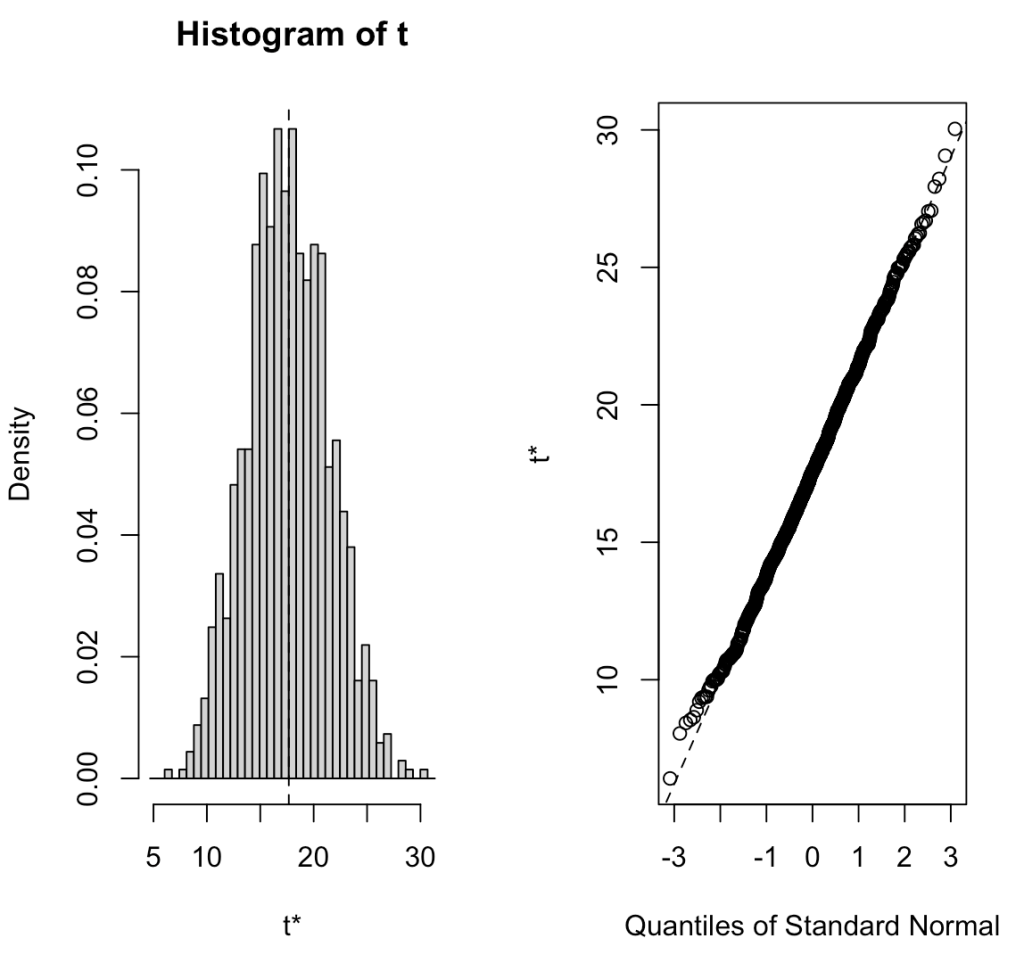
R offers a bootstrap function from the boot package that allows you to do the same thing without writing out your own for() loop. Below is an example of coding the same procedure in the boot package and the outputs the function provides, which are similar to the output we get above, save for slight differences due to random sampling.
# write a function to calculate the mean of our sample data
sample_mean <- function(x, d){
return(mean(x[d]))
}
# run the boot() function
library(boot)
# run the boot function
boot_func_output <- boot(dat, statistic = sample_mean, R = 1000)
# produce a plot of the output
plot(boot_func_output)
# get the mean and standard error
boot_func_output
# get 95% CI around the mean
boot.ci(boot_func_output, type = "basic", conf = 0.95)
We can bootstrap pretty much anything we want. We don’t have to limit ourselves to producing the distribution around the mean of a population. For example, let’s bootstrap regression coefficients to understand the uncertainty in them.
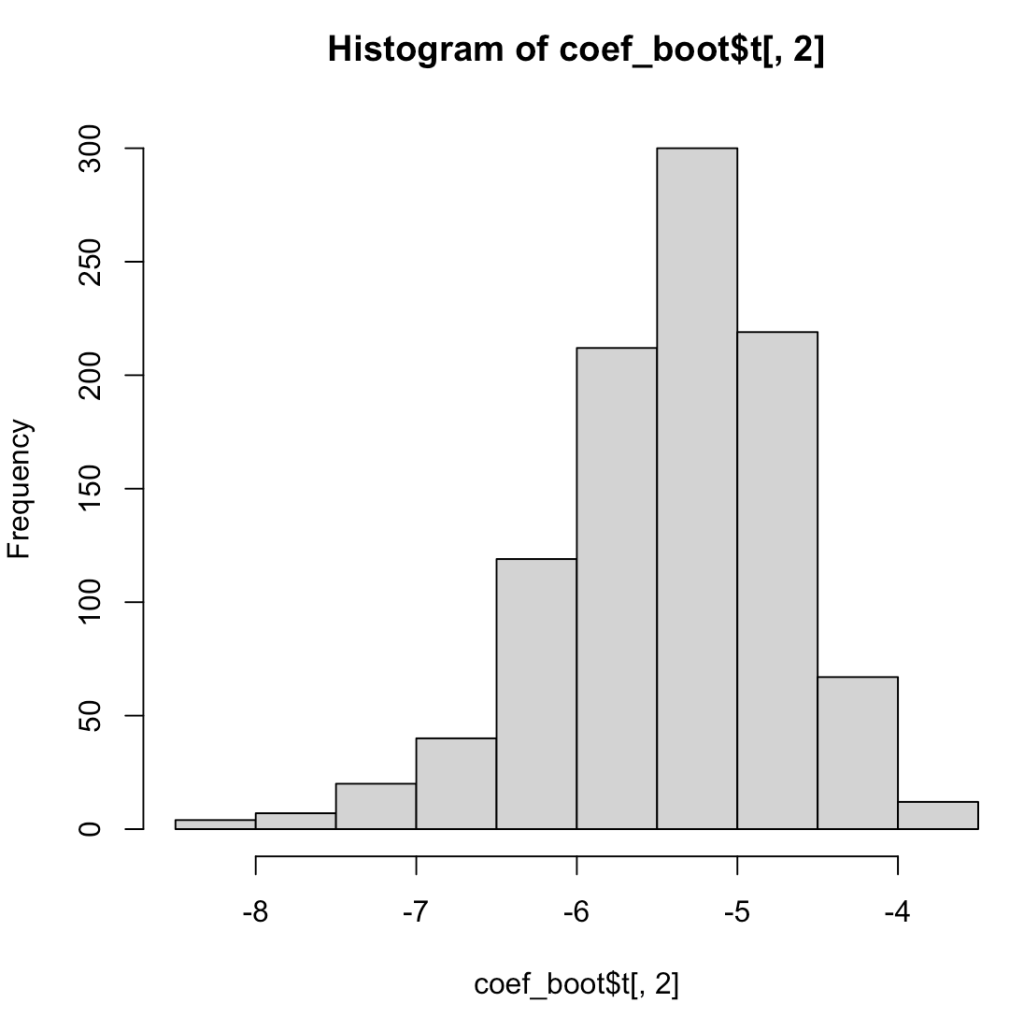
First, let’s use the boot() function to conduct our analysis. We will fit a simple linear regression predicting miles per gallon from engine weight using the mtcars package. We will then write a function for that uses a random sample of rows to create the same linear regression and store those coefficients from 1000 linear regressions so that we can plot a histogram representing the slope coefficient from the resampled models and also summarize the distribution with confidence intervals.
# load the mtcars data
d <- mtcars d %>%
head()
# fit a regression model
fit_mpg <- lm(mpg ~ wt, data = d)
summary(fit_mpg)
coef(fit_mpg)
confint(fit_mpg)
# Write a function that can perform a bootstrap over the intercept and slope of the model
# bootstrap function
reg_coef_boot <- function(data, row_id){
# we want to resample the rows
fit <- lm(mpg ~ wt, data = d[row_id, ])
coef(fit)
}
# run this once on a small subset of the row ids to see how it works
reg_coef_boot(data = d, row_id = 1:20)
# run the boot() function 1000 times
coef_boot <- boot(data = d, reg_coef_boot, 1000)
# check the output (coefficient and SE)
coef_boot
# get the confidence intervals
boot.ci(coef_boot, index= 2)
# all 1000 of the bootstrap resamples can be called coef_boot$t %>%
head()
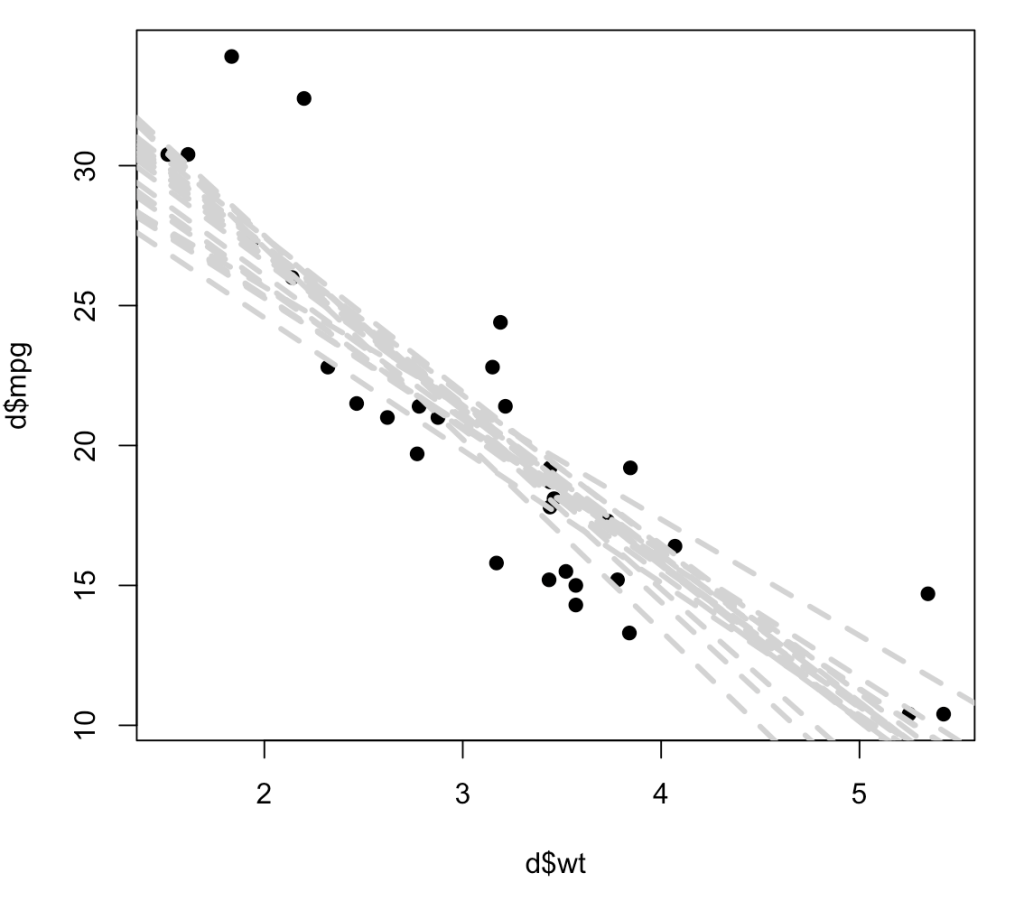
# plot the first 20 bootstrapped intercepts and slopes over the original data
plot(x = d$wt,
y = d$mpg,
pch = 19)
for(i in 1:20){
abline(a = coef_boot$t[i, 1],
b = coef_boot$t[i, 2],
lty = 2,
lwd = 3,
col = "light grey")
}
## histogram of the slope coefficient
hist(coef_boot$t[, 2])
We can do this by hand if we don’t want to use the built in boot() function. (SIDE NOTE: I usually prefer to code my own resampling and simulations as it gives me more flexibility with respect to the things I’d like to add or the values I’d like to store from each iteration.
Below, instead of producing a histogram of just the slope coefficient, I use both the resampled intercept and slope and add 20 (of the 1000) lines to a scatter plot to show the way in which each of these lines represents a plausible regression line for the data. As you can see, the regression line confidence interval is starting to take shape, even with just 20 out of 1000 resamples, and this gives us a good understanding of not only the variability in our possible regression line fit for the underlying data but also, perhaps should make us less overconfident in our research findings knowing that there are many possible outcomes from the sample data we have obtained.
## 1000 resamples
n_samples <- 1000
## N observations
n_obs <- nrow(mtcars)
## empty storage data frame for the coefficients
coef_storage <- data.frame(
intercept = rep(NA, n_samples),
slope = rep(NA, n_samples)
)
for(i in 1:n_samples){
## sample dependent and independent variables
row_ids <- sample(1:n_obs, size = n_obs, replace = TRUE)
new_df <- d[row_ids, ]
## construct model
model <- lm(mpg ~ wt, data = new_df)
## store coefficients
# intercept
coef_storage[i, 1] <- coef(model)[1]
# slope
coef_storage[i, 2] <- coef(model)[2]
}
## see results
head(coef_storage)
tail(coef_storage)
## Compare the results to those of the boot function
apply(X = coef_boot$t, MARGIN = 2, FUN = mean)
apply(X = coef_storage, MARGIN = 2, FUN = mean)
apply(X = coef_boot$t, MARGIN = 2, FUN = sd)
apply(X = coef_storage, MARGIN = 2, FUN = sd)
## plot first 20 lines
plot(x = d$wt,
y = d$mpg,
pch = 19)
for(i in 1:20){
abline(a = coef_storage[i, 1],
b = coef_storage[i, 2],
lty = 2,
lwd = 3,
col = "light grey")
}
Simulating a relationship between two variables
As discussed in Part 1, simulation differs from resampling in that we use the parameters of the observed data to compute a new distribution, versus sampling from the data we have on hand.
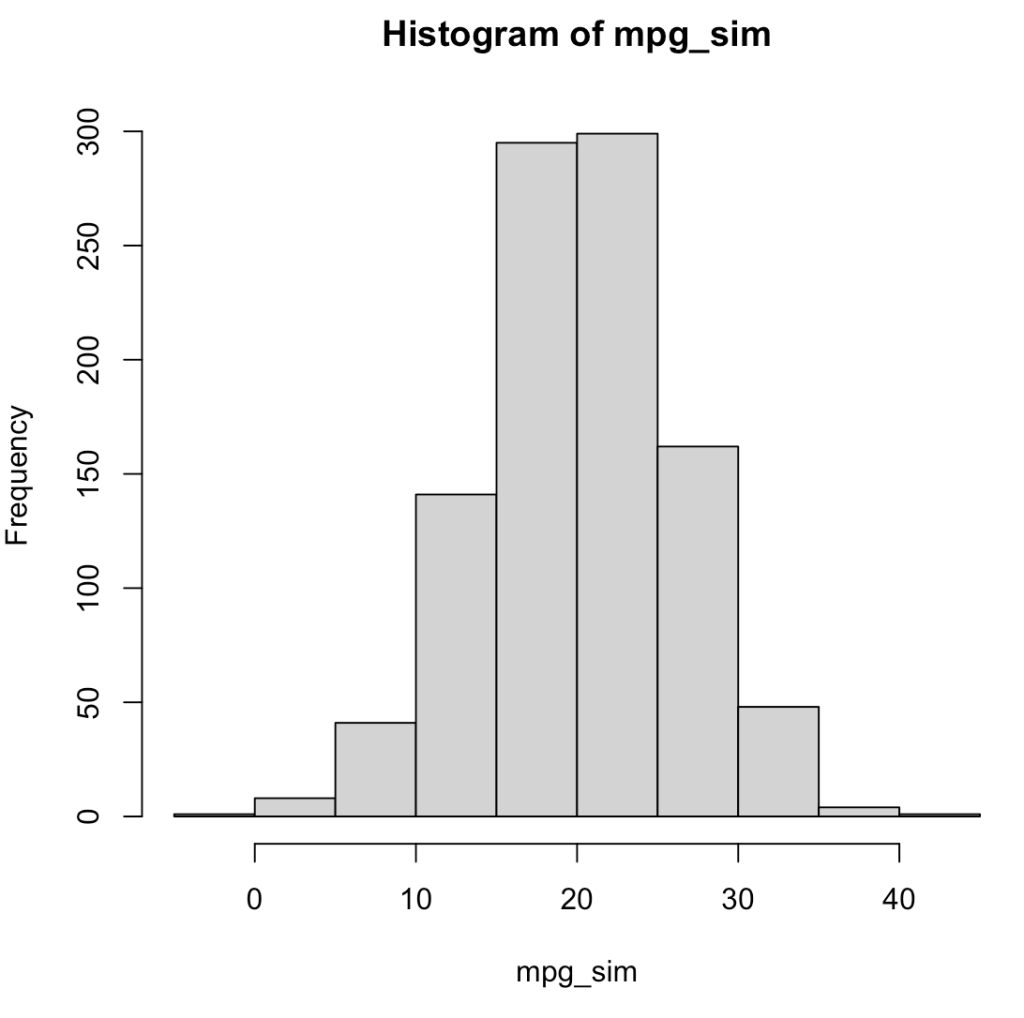
For example, using the mean and standard deviation of mpg from the mtcars data set, we can simulate 1000 random draws from a normal distribution.
## load the mtcars data set d <- mtcars ## make a random draw from the normal distribution for mph set.seed(5) mpg_sim <- rnorm(n = 1000, mean = mean(d$mpg), sd = sd(d$mpg)) ## plot and summarize hist(mpg_sim) mean(mpg_sim) sd(mpg_sim)
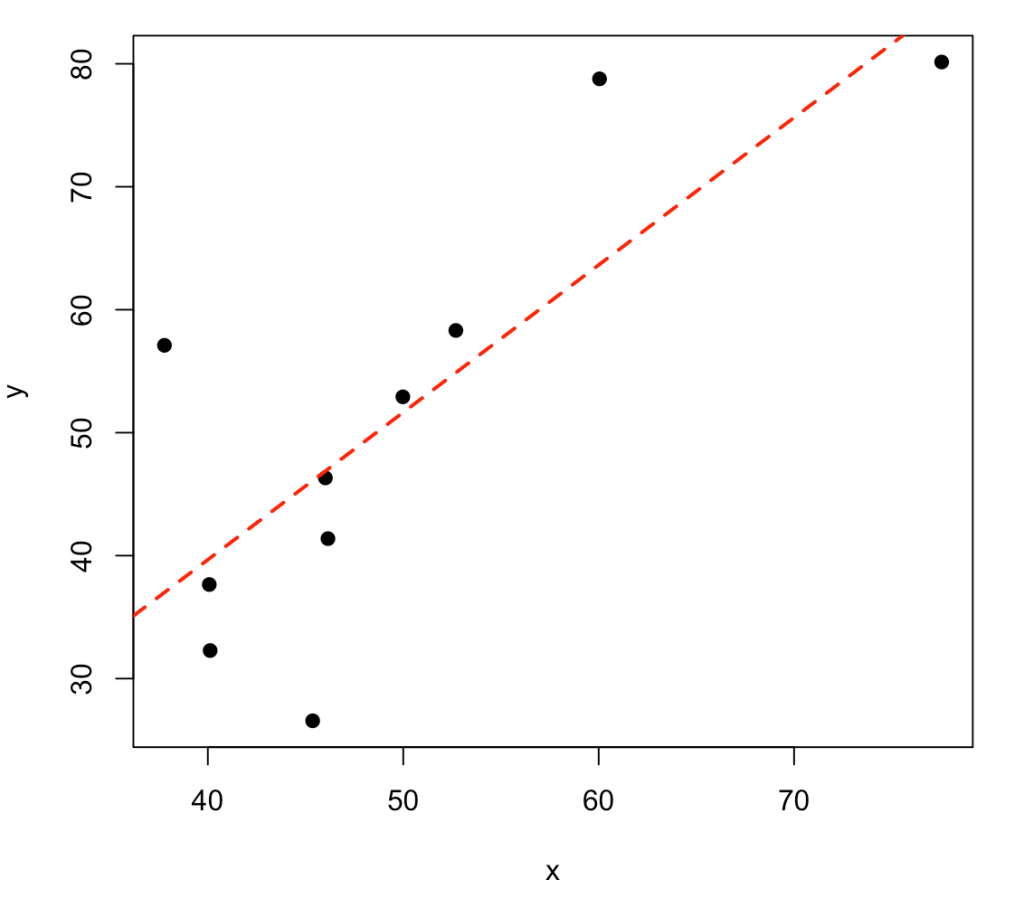
Frequently, we are interested in the relationship between two variables (e.g., correlation, regression, etc.). Let’s simulate two variables, x and y, which are linearly related in some way. To do this, we first simulate the variable x and then simulate y to be x plus some level of random noise.
# simulate x and y set.seed(1098) x <- rnorm(n = 10, mean = 50, sd = 10) y <- x + rnorm(n = length(x), mean = 0, sd = 10) # put the results in a data frame dat <- data.frame(x, y) dat # how correlated are the two variables cor.test(x, y) # fit a regression for the two variables fit <- lm(y ~ x) summary(fit) # plot the two variables with the regression line plot(x, y, pch = 19) abline(fit, col = "red", lwd = 2, lty = 2)
Simulating a data set with multiple variables
Frequently, we might have a hypothesis regarding how correlated multiple variables are with each other. The example above produced a relationship of two variables with a direct relationship between them along with some noise. We might want to specify this relationship given a correlation coefficient or covariance between them. Additionally, we might have more than two variables that we want to simulate relationships between.
To do this in R we can take advantage of two packages:
- MASS via the mvrnorm()
- mvtnorm via the mvrnorm()
Both packages have a function for simulating multivariate normal distributions. The primary difference is that the Sigma argument in the MASS package function, mvrnorm(), accepts a covariance matrix while the sigma argument in the mvtnorm package, rmvnorm() accepts a correlation matrix. I’ll show both examples but I tend to stick with the mtvnorm package because (at least for my brain) it is easier for me to think in terms of correlation coefficients instead of covariances.
First we simulate some data:
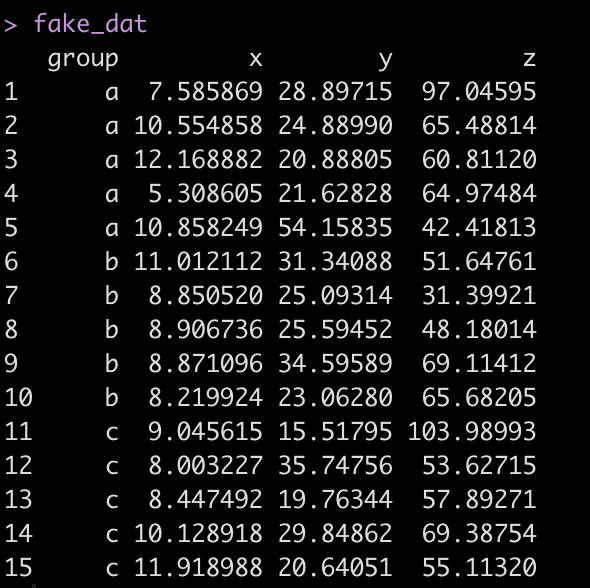
## create fake data
set.seed(1234)
fake_dat <- data.frame(
group = rep(c("a", "b", "c"), each = 5),
x = rnorm(n = 15, mean = 10, sd = 2),
y = rnorm(n = 15, mean = 30, sd = 10),
z = rnorm(n = 15, mean = 75, sd = 20)
)
fake_dat
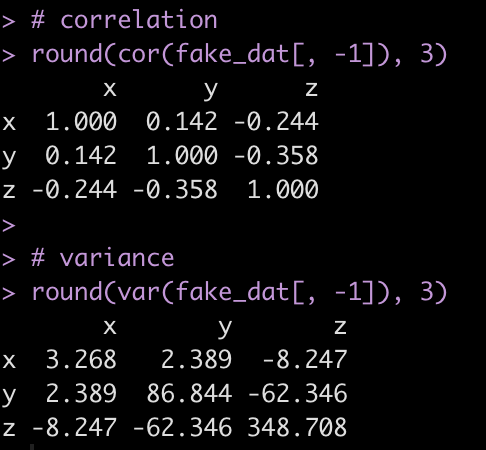
Look at the correlation and variance between the three numeric variables.
# correlation round(cor(fake_dat[, -1]), 3) # variance round(var(fake_dat[, -1]), 3)
We can use this information to simulate new x, y, or z variables.
Simulating x and y with the MASS package
Remember, for the MASS package, the Sigma argument is a matrix of covariances for the variables you are simulating from a multivariate normal distribution.
## get a vector of the mean for each variable
variable_means <- apply(X = fake_dat[, c("x", "y")], MARGIN = 2, FUN = mean)
## Get a matrix of the covariance between x and y
variable_sigmas <- var(fake_dat[, c("x", "y")])
## simulate 1000 new x and y variables using the MASS package
set.seed(98)
new_sim <- MASS::mvrnorm(n = 1000, mu = variable_means, Sigma = variable_sigmas)
head(new_sim)
### look at the results relative to the original x and y
## column means
variable_means
apply(X = new_sim, MARGIN = 2, FUN = mean)
## covariance
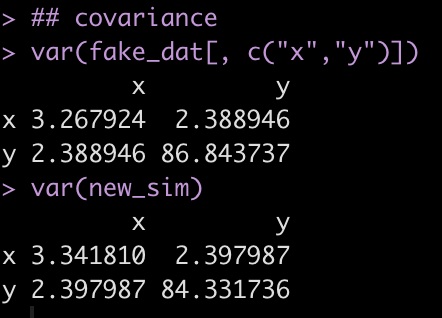
var(fake_dat[, c("x","y")])
var(new_sim)
Notice that the variance between x and y (the off diagonal of the matrix) is very similar between the fake data (the observed sample) and the simulated data (new_sim).
Simulating x and y with the mtvnorm package
Different than the MASS package, The rmvnorm() function from the mtvnorm package requires the sigma argument to be a correlations matrix.
Let’s repeat the above process with our fake_dat and simulate a relationship between x and y.
## get a vector of the mean for each variable
variable_means <- apply(X = fake_dat[, c("x", "y")], MARGIN = 2, FUN = mean)
## Get a matrix of the correlation between x and y
variable_cor <- cor(fake_dat[, c("x", "y")])
## simulate 1000 new x and y variables using the mvtnorm package
set.seed(98)
new_sim <- mvtnorm::rmvnorm(n = 1000, mean = variable_means, sigma = variable_cor)
head(new_sim)
### look at the results relative to the original x and y
## column means
variable_means
apply(X = new_sim, MARGIN = 2, FUN = mean)
## correlation
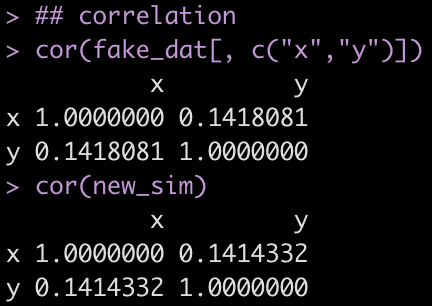
cor(fake_dat[, c("x","y")])
cor(new_sim)
Similar to the previous example, we notice that the off diagonal correlation coefficient between x and y is very similar when comparing the simulated data to the fake data.
So, what is happening here? Both packages produce the same result, one uses a covariance matrix and the other uses a correlation matrix. The kicker here is understanding the relationship between covariance and correlation. Covariance is explaining how two variables vary together, however, its units aren’t on a scale that is directly interpretable to us. But, we can convert the covariance between two variables to a correlation by dividing their covariance by the product of their individual standard deviations.
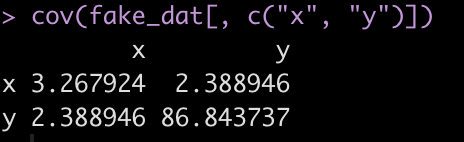
For example, here is the covariance matrix between x and y in the fake data set.
cov(fake_dat[, c("x", "y")])
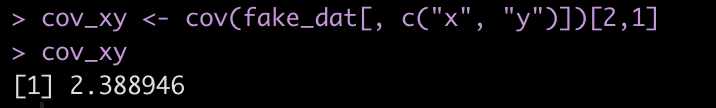
The covariance between the two variables is on the off diagonal, 2.389. We can store this in its own element.
cov_xy <- cov(fake_dat[, c("x", "y")])[2,1]
cov_xy
Let’s store the standard deviation of both `x` and `y` in their own elements to make the equation easier to read.
sd_x <- sd(fake_dat$x) sd_y <- sd(fake_dat$y)
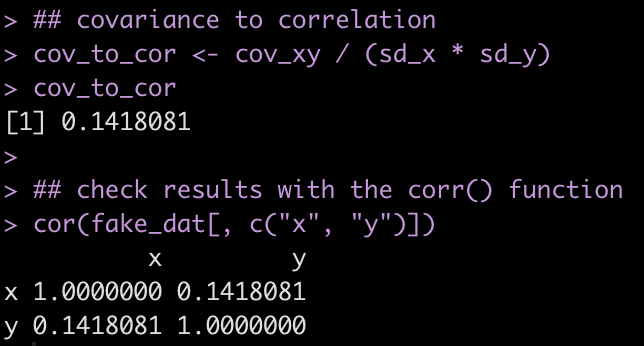
Finally, we calculate the correlation by dividing the covariance by the product of the two standard deviations and check our results by calling the cor() function on the two variables.
## covariance to correlation
cov_to_cor <- cov_xy / (sd_x * sd_y)
cov_to_cor
## check results with the corr() function
cor(fake_dat[, c("x", "y")])
By dividing the covariance by the product of the standard deviation of the two variables we can see the relationship between covariance and correlation and now understand why the results from the MASS and mtvnorm produce similar results. Understanding this relationship becomes valuable when we move onto simulating more complex relationships, for example when simulating mixed models.
What about three variables?
What if we want to simulate all three variables — x, y, and z?
All we need is a larger covariance or correlation matrix, depending on which of the above packages you’d like to use. Since we usually won’t be creating these matrices from a data set, as I did above, I’ll show how to create your own matrix and run the simulation.
First, let’s store a vector of plausible mean values for x, y, and z.
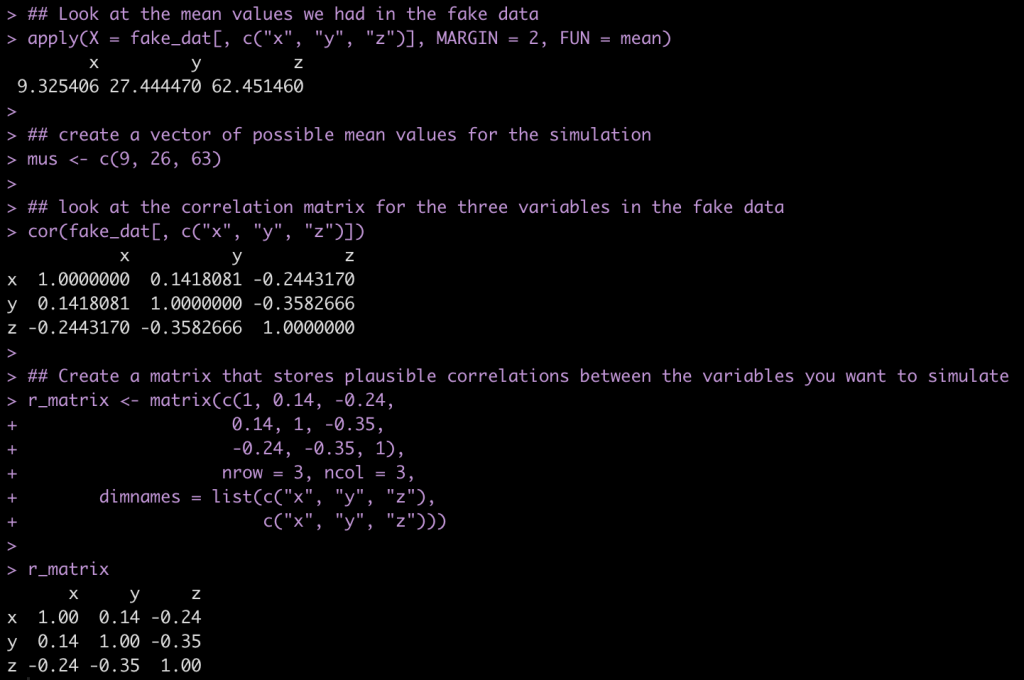
## Look at the mean values we had in the fake data
apply(X = fake_dat[, c("x", "y", "z")], MARGIN = 2, FUN = mean)
## create a vector of possible mean values for the simulation
mus <- c(9, 26, 63)
## look at the correlation matrix for the three variables in the fake data
cor(fake_dat[, c("x", "y", "z")])
## Create a matrix that stores plausible correlations between the variables you want to simulate
r_matrix <- matrix(c(1, 0.14, -0.24,
0.14, 1, -0.35,
-0.24, -0.35, 1),
nrow = 3, ncol = 3,
dimnames = list(c("x", "y", "z"),
c("x", "y", "z")))
r_matrix
Next, we create 1000 simulations of a multivariate normal distribution between x, y, and z. We then compare our correlation coefficients between the fake data, which is our observed sample, and the simulated data
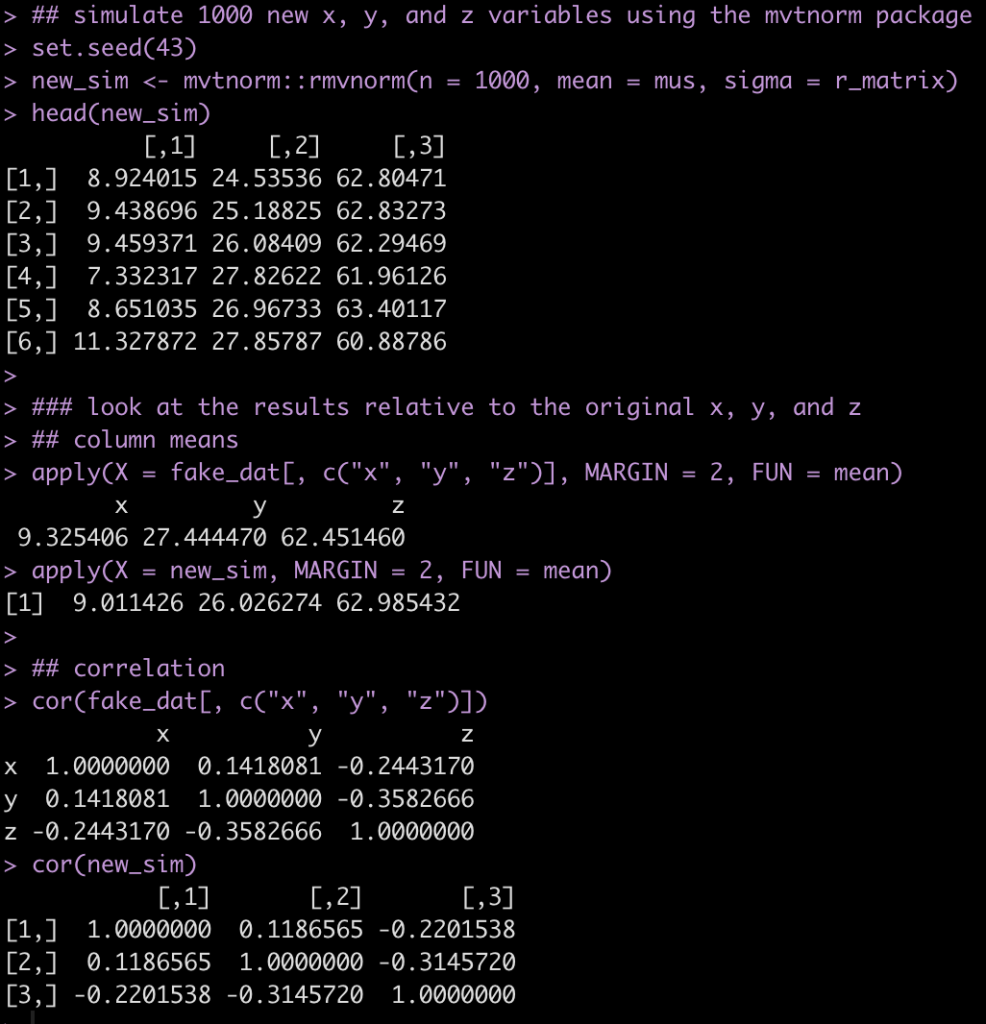
## simulate 1000 new x, y, and z variables using the mvtnorm package
set.seed(43)
new_sim <- mvtnorm::rmvnorm(n = 1000, mean = mus, sigma = r_matrix)
head(new_sim)
### look at the results relative to the original x, y, and z
## column means
apply(X = fake_dat[, c("x", "y", "z")], MARGIN = 2, FUN = mean)
apply(X = new_sim, MARGIN = 2, FUN = mean)
## correlation
cor(fake_dat[, c("x", "y", "z")])
cor(new_sim)
The results from the simulation are pretty similar to the fake_dat dataset. If you recall from the correlation matrix and the vector of means, we didn’t use exact values from the observed data, so that, along with the random draws from the multivariate normal distribution, leads to the small amount of differences.
Wrapping Up
In this second installment of the Simulations in R series we’ve walked through how to code our own bootstrap resampling for both means and regression coefficients. We then progressed to building simulations of both bivariate and multivariate normal distributions. This, along with the basic info in Part 1, will serve us well as we progress forward in our work and begin to explore using simulations for comparisons between group means (simulated t-tests) and building regression models.
As always, all of the code is available in the Github repository.