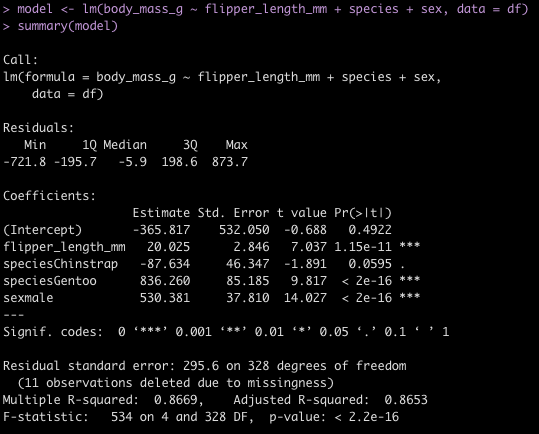
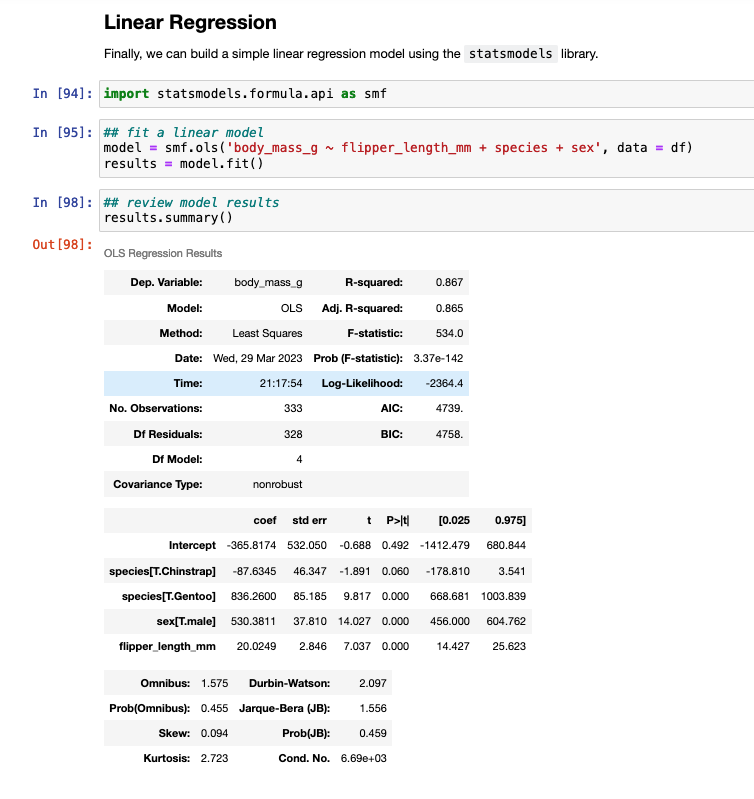
I’ve discussed using loops for a number of statistical tasks (simulation, optimization, Gibbs sampling) as well as data processing tasks, such as writing data outputs to separate excel tabs within one excel file and creating a multiple page PDF with a plot on each page.
Today, I want to expand the loop function to produce separate SVG file plots and have R save those directly to a folder stored on my computer. The goal here is to have the separate plots in one place so that I can upload those files directly to a web app and allow them to be viewable for a decision-maker.
NOTE: You can save these files in other formats (e.g., jpeg, png). I chose SVG because it was the primary file type I had been working with.
Data
To keep the example simple, we will be using the {mtcars} data set, which is freely available in R. I’m going to set the cylinder (cyl) variable to be a factor as that is the variable that we will build our separate plot files for. In the sport setting, you can think of this as player names or player IDs, where you are building a plot for each individual, looping over them and producing a separate plot file.
library(tidyverse)
library(patchwork)
theme_set(theme_bw())
## data
dat <- mtcars %>%
mutate(cyl = as.factor(cyl))
Example Plots
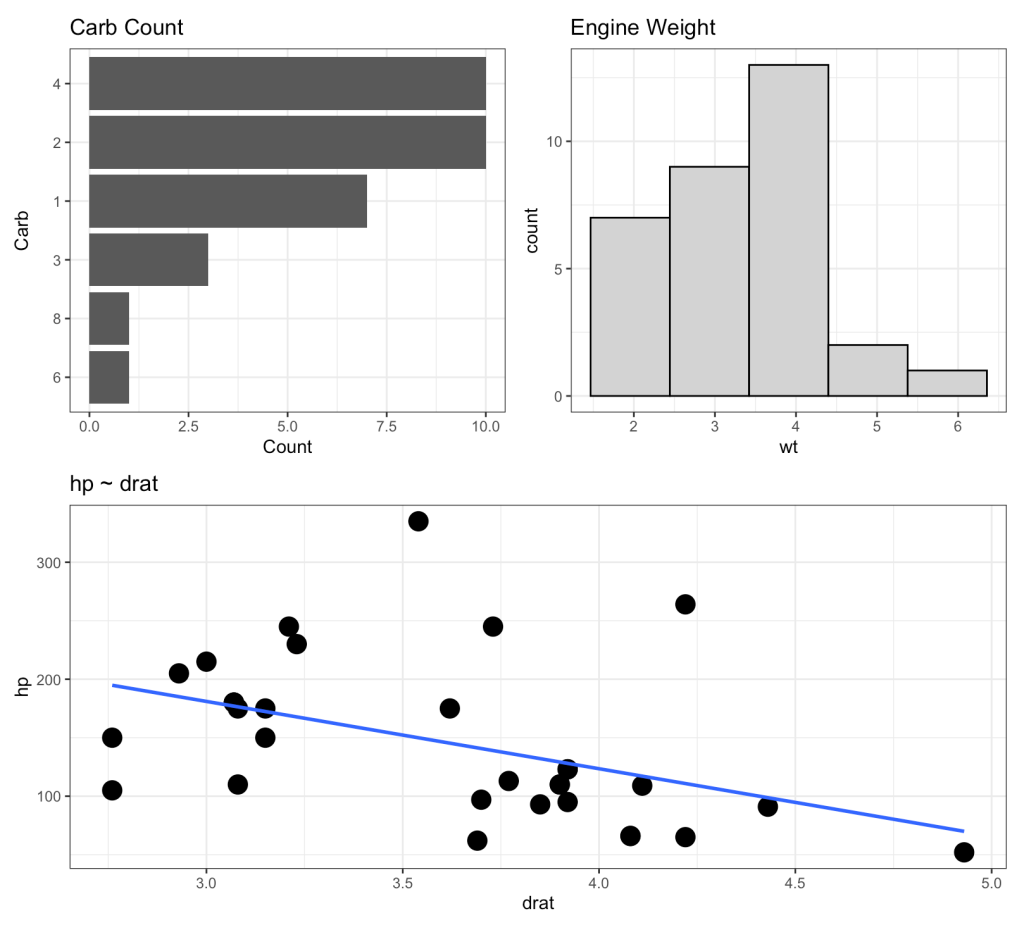
Here is an example of the three types of plots we will build. We will wrap the three plots together using the {patchwork} package. The below plot is using all of the data but our goal will be to produce a loop function that creates the same plot layout using data for each of the three cylinder types.
p1 <- dat %>%
ggplot(aes(x = drat, y = hp)) +
geom_point(size = 5) +
geom_smooth(method = "lm",
se = FALSE) +
ggtitle("hp ~ drat")
p2 <- dat %>%
count(carb) %>%
mutate(carb = as.factor(carb)) %>%
ggplot(aes(x = n, y = reorder(carb, n))) +
geom_col() +
labs(x = "Count",
y = "Carb",
title = "Carb Count")
p3 <- dat %>%
ggplot(aes(x = wt)) +
geom_histogram(fill = "light grey",
color = "black",
bins = 5) +
ggtitle("Engine Weight")
(p2 | p3) / p1
Creating the loop for plotting
First, we create a function that produces the plots above. Basically, I’m taking the plotting code from above and wrapping it in a function. The function takes in input, i, and runs through the three plots for that input, at the end using the ggsave() function to save each plot to the dedicated file path.
# create a plot function for each cyl
plt_func <- function(i){
p1 <- i %>%
ggplot(aes(x = drat, y = hp)) +
geom_point(size = 5) +
geom_smooth(method = "lm",
se = FALSE) +
ggtitle("hp ~ drat")
p2 <- i %>%
count(carb) %>%
mutate(carb = as.factor(carb)) %>%
ggplot(aes(x = n, y = reorder(carb, n))) +
geom_col() +
labs(x = "Count",
y = "Carb",
title = "Carb Count")
p3 <- i %>%
ggplot(aes(x = wt)) +
geom_histogram(fill = "light grey",
color = "black",
bins = 5) +
ggtitle("Engine Weight")
three_plt <- (p2 | p3) / p1
ggsave(three_plt, file = paste0(unique(i$cyl), ".svg"))
}
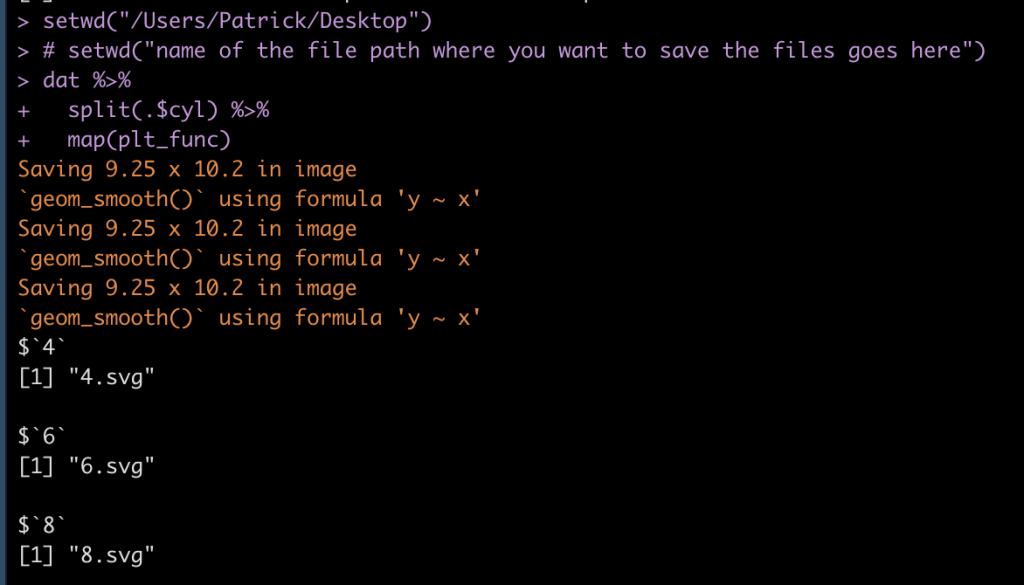
Then, we use the split() function to split the data frame into a named list with each cylinder type being it’s own list that contains a data frame. The map() function then creates the loop over that list and for each element of the list (for each cylinder type) it runs our plot function above and saves the results. Notice that I’ve specified setwd() to indicate where I want the files to be saved to. If you are saving thousands of files at once and you don’t specify this and have your working directory defaulted to your desktop, it becomes a mess pretty quick (trust me!).
# setwd("name of the file path where you want to save the files goes here")
dat %>%
split(.$cyl) %>%
map(plt_func)
Once you’ve run the loop, your R output should look like this, where we see that each list element (cylinder) is being saved as an SVG file.
Our folder has the plot outputs:
If I click on any one of the SVG files I get the desired plot.
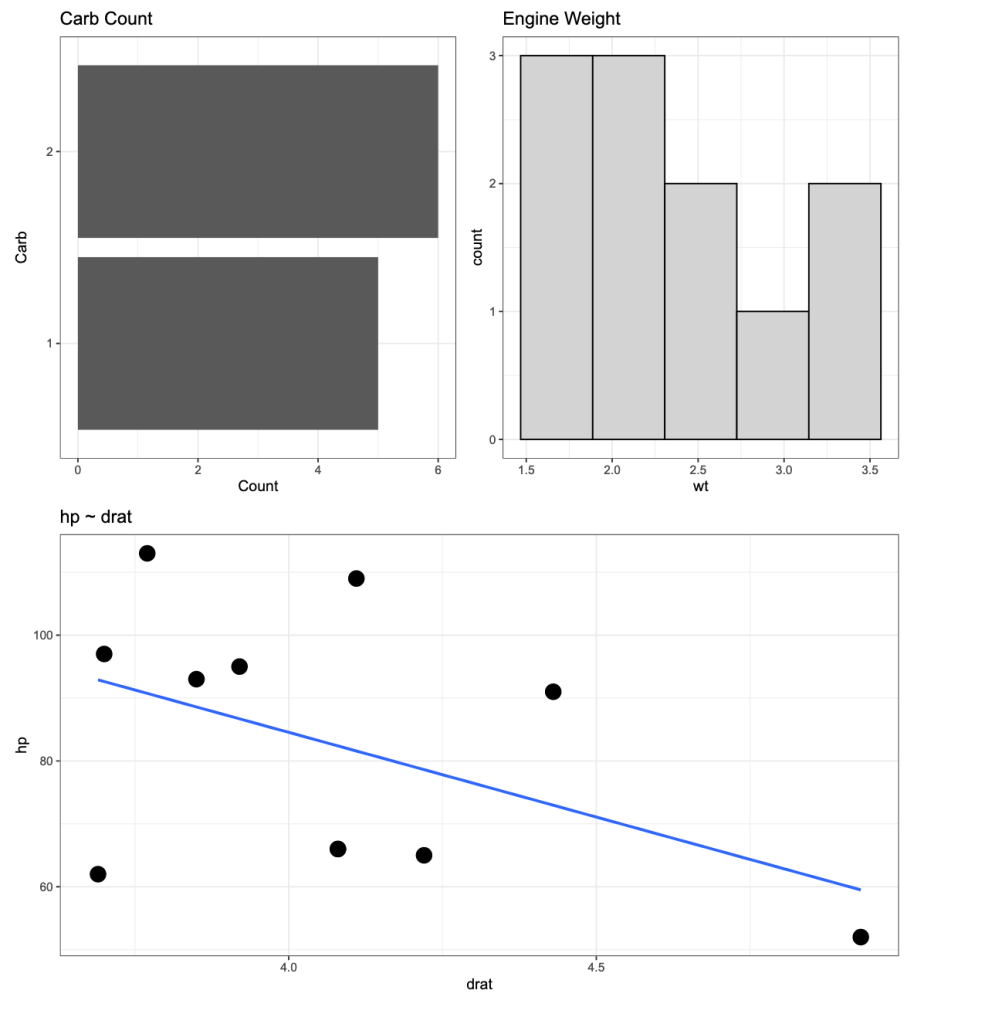
The above is for a 4 cylinder vehicle. Notice that I didn’t specify this at the top of the plot because my initial assumption was that I would be uploading the individual SVG files to a web application where there is a webpage dedicated to each cylinder type. Therefore, naming the plots by cylinder type would be redundant. However, if you want to add a plot to the {pathwork} layout about, you can use the plot_annotation() function.
(p2 | p3) / p1 + plot_annotation(title = "Engine cylinders")
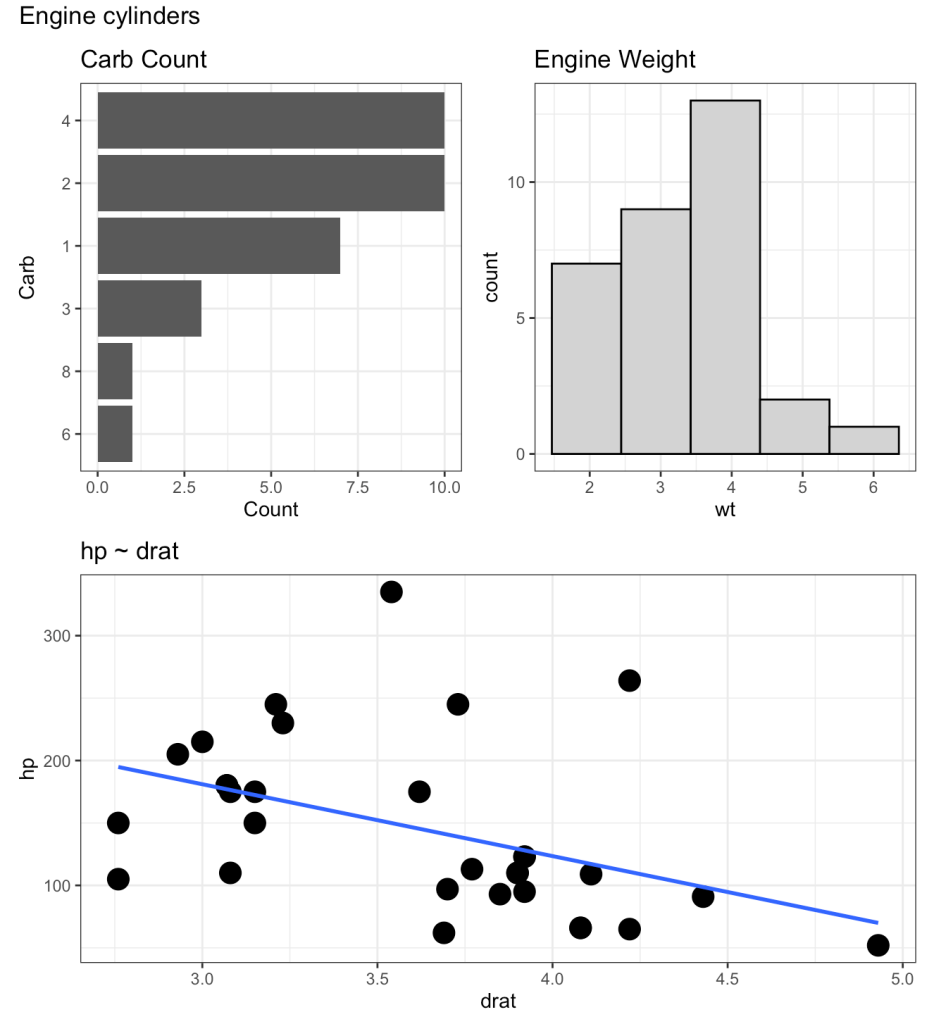
We can add the plot_annotation() function to the loop but instead of a generic title, like above, we will need to create a bespoke title within the loop that stores each cylinder type. To do this, we use the paste() function to add the cylinder number in front of the word “cylinder” in our plot title name.
plt_func <- function(i){
p1 <- i %>%
ggplot(aes(x = drat, y = hp)) +
geom_point(size = 5) +
geom_smooth(method = "lm",
se = FALSE) +
ggtitle("hp ~ drat")
p2 <- i %>%
count(carb) %>%
mutate(carb = as.factor(carb)) %>%
ggplot(aes(x = n, y = reorder(carb, n))) +
geom_col() +
labs(x = "Count",
y = "Carb",
title = "Carb Count")
p3 <- i %>%
ggplot(aes(x = wt)) +
geom_histogram(fill = "light grey",
color = "black",
bins = 5) +
ggtitle("Engine Weight")
cyl_name <- i %>%
select(cyl) %>%
distinct(cyl) %>%
pull(cyl)
three_plt <- (p2 | p3) / p1 + plot_annotation(title = paste(cyl_name, "cylinder", sep = " ")) ggsave(three_plt, file = paste0(unique(i$cyl), ".svg")) } # setwd("name of the file path where you want to save the files goes here") dat %>%
split(.$cyl) %>%
map(plt_func)
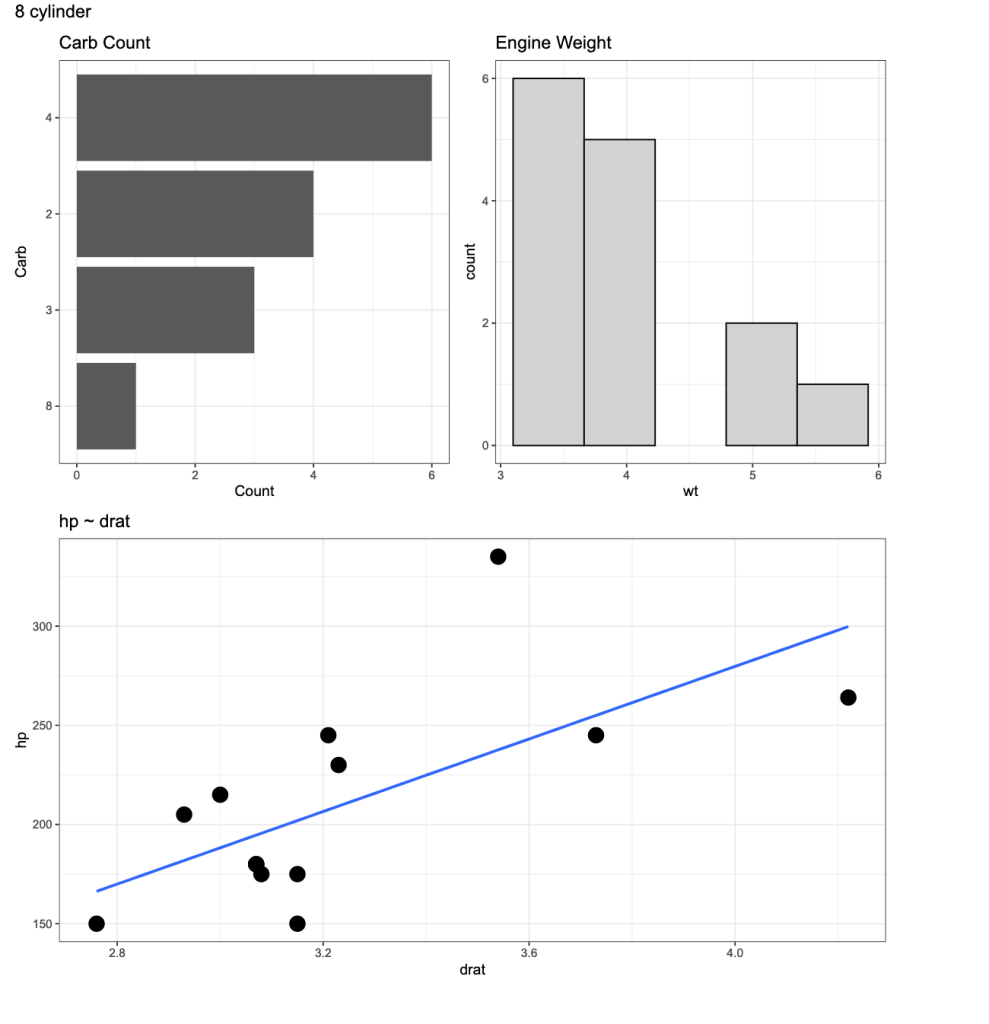
Now we have plots with named titles.
Wrapping Up
During those times where you need to produce several individual plots, rather than doing them one-by-one, leverage R’s loop functions to rapidly produce multiple plots in one shot.
The full code is accessible on my GITHUB page.