Previously I’ve used {rstanarm}, {brms}, and Stan for fitting Bayesian models. However, as I continue to work on improving my Python skills, I figured I’d try and delve into the PyMC3 framework for fitting such models. This article will go through the following steps:
- Fitting the model
- Making a point estimate prediction
- Making a point estimate prediction with uncertainty
- Calculating a posterior predictive distribution
I’ve covered the last three steps in a prior blog on making predictions with a Bayesian model. I know there are probably functions available in PyMC3 that can do these things automatically (just as there are in {rstanarm}) but instead of falling back on those, I create the posterior distributions here using numpy and build them myself.
The entire code and data are available on my GITHUB page, where I also have the model coded in {rstanarm}, for anyone interested in seeing the steps in a different code language.
Loading Libraries & Data
The data I’ll be using is the {mtcars} data set, which is available in R. I’ve saved a copy in .csv format so that I can load it into my Jupyter notebook.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pymc3 as pm
import arviz as az
import os
# load mtcars
d = pd.read_csv('mtcars.csv')
d.head()
Exploratory Data Analysis
The model will regress mpg on engine weight (wt). Let’s plot and describe those two variables so that we have a sense for what we might be working with.
Linear Regression
Before fitting the Bayesian model, I want to fit a simple regression model to see what the coefficients look like.
import statsmodels.api as sm x = d['wt'] y = d['mpg'] x = sm.add_constant(x) fit = sm.OLS(y, x).fit() fit.summary()
We can see that for every one unit increase in engine weight the miles per gallon decrease, on average, by about 5.3.
Bayesian Regression (PyMC3)
Fitting a Bayesian regression model in PyMC3 requires us to specify some priors. For this model I’ll use a prior intercept of 40 ± 10 and a prior beta for the wt variable of 0 ± 10. The thing to note here is that the priors I’m specifying priors were created by first looking at the data that has been collected (which is technically cheating). Normally we would have priors BEFORE collecting our data (using prior published research, data from a pilot study, prior intuition, etc) and then combine the prior with the observations to obtain a posterior distribution. However, the aim here is to understand how to code the model, so I’ll use these priors. I’ll write a longer blog on priors and what they do to a model in the coming weeks.
Some notes on fitting the model in PyMC3:
- The model is named ‘fit_b‘
- We specify the intercept as variable ‘a‘
- The beta coefficient for wt is called ‘b‘
- Both the intercept and slope re fit with normally distributed priors
- Finally, ‘e‘ represents the model error and it is fit with a a Half Cauchy prior
- Once the priors are set, the model is specified (y_pred) as mu = a + b * wt + e
- The trace_b object stores our posterior samples, 2000 of them of which the first 1000 will be discarded because they are there to allow the model to tune itself
, sd = e, observed = d['mpg'])
trace_b = pm.sample(2000, tune = 1000)
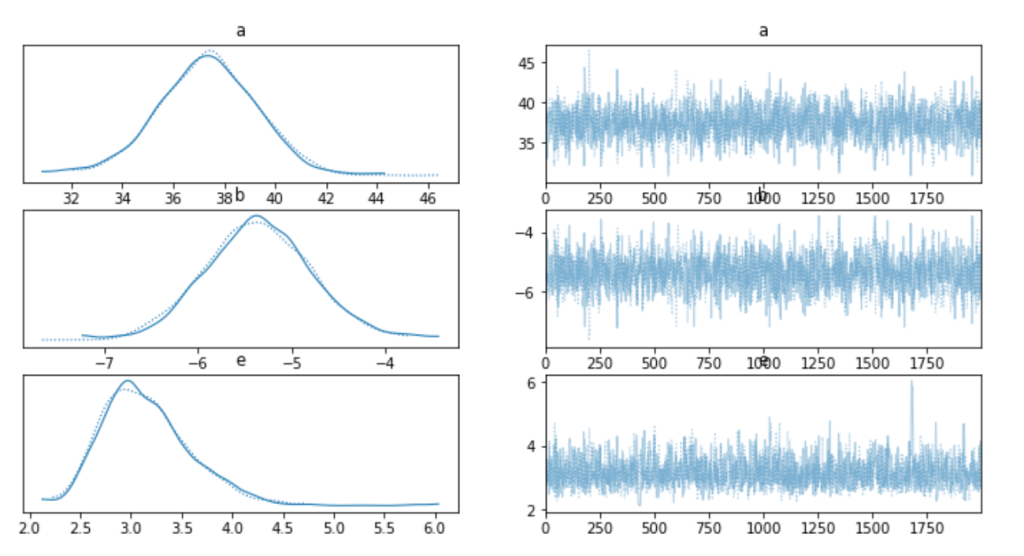
Once the model has been fit we plot the trace plots to see how well it performed.
We can also directly call the mean and standard deviation values of our fitted model, which are relatively similar to what we saw with the linear regression model above.
Point Predictions
Next, we want to make a single point prediction for the mpg would expect, on average, when wt is a specific value (in this example we will use wt = 3.3).
To do this, we simply store the average value of the posterior coefficients from our Bayesian regression and apply the specified model:
mu = a + b * new_wt
A car with an engine weight of 3.3 would get, on average, 19.7 mpg.
Point Prediction with Uncertainty
The point estimate is interesting (I guess), but there is uncertainty around that estimate as point predictions are never exact. We can compliment this point estimate by unveiling the uncertainty around it. The point prediction ± uncertainty interval informs us of the average value of mpg along with the uncertainty of the coefficients in our model.
To do this, we create a random sample of 1000 values from the posterior distributions for our model intercept and beta coefficient. Each of these 1000 values represent a potential intercept and slope that would be consistent with our data, which shows us the uncertainty that we have in our estimates. When we use the model equation, multiplying each of these 1000 values by the new_wt value we obtain 1000 possible predicted values of mpg given a weight of 3.3.
With this posterior distribution we can then plot a histogram of the results and obtain summary statistics such as the mean, standard deviation, and 90% credible interval.
Posterior Predictive Distribution
Finally, instead of just knowing the average predicted value of mpg ± uncertainty for the population, we might be interested in knowing what the predicted value of mpg would be for a new car in the population with a wt of 3.3. For that, we calculate the posterior predictive distribution. The uncertainty in this predictive distribution will be larger than the point prediction with uncertainty because we are using the posterior model error added to our prediction.
First, similar to the model coefficients, we have to get the random draws of our error term, which we will call sigma.
Next, we run the model as we did in step 2 above; however, we also add to each of the 1000 posterior predictions the sigma value by taking a random draw from a normal distribution with a mean of 0 and standard deviation equal to the sigma sample values.
pred_dist = intercept_sample + beta_sample * new_wt_rep + np.random.normal(loc = 0, scale = sigma_sample, size = 1000)
Finally, we plot the distribution of our predicted values along with the mean, standard deviation, and 90% credible interval. Notice that these values are larger than what we obtained in step 2 because we are now taking into account additional uncertainty about the new wt observation.
Wrapping Up
That’s a brief intro to Bayesian regression with PyMC3. There are a lot more things that we can do with PyMC3 and it’s available functions. My goal is to put together more blog articles on Bayesian modeling with both R and Python so show their flexibility. If you spot any errors, please let me know.
The data and full code (along with a companion code in {rstanarm}) is available on my GITHUB page.