One of the most frustrating aspects of building {shiny} apps is dealing with columns that have NAs when outputting tables. This is common in sport when dealing with players from different position groups who may have different stats that describe performance for those positions. Rather than writing a long series of if/else statements, I prefer to streamline the process by dropping those columns prior to returning the table of data. Not only does this make the app run smoothly but it also is easier to debug or add additional table information without having to deal with a lot of nested if/else statements.
The full code is accessible on my GITHUB page.
Load Packages & Simulate Data
## Removing columns with NA for fluid table building in shiny ## packages --------------------------------------------------- library(tidyverse) library(shiny) ## simulate data ---------------------------------------------- d <- tribble( ~player, ~position, ~stat1, ~stat2, ~stat3, 'Frank', 'Pitcher', 10, NA, 33, 'Tom', 'Batter', NA, 14, 12, 'Jeff', 'Batter', NA, 5, NA, 'Harold', 'Pitcher/Batter', 12, 33, 9 ) d
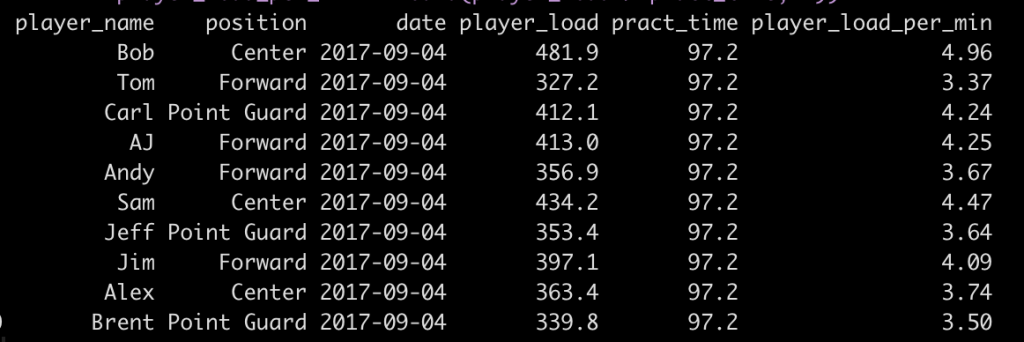
We can see from our little data set that different players have different stats populated. We really don’t want our users to deal with having to see NA in the table output. So, we need to devise a way to drop the columns with NA’s once a specific player has been selected.
Dropping Columns with NA in Base R
Let’s select on player and attempt to drop their columns with NA. In base R we will use the colSums() function to produce a count of the number of NA’s in each column.
## remove columns with NA for Frank, using Base R -------------------------- frank <- d %>% filter(player == 'Frank') # colSums() can be used to count the NA's in each column colSums(is.na(frank))
We can see that stat2 has 1 NA while the other 4 columns are complete. We can use this information to retain those four columns and drop stat2.
frank[ , colSums(is.na(frank)) == 0]
Dropping Columns with NA in {dplyr}
We can perform a similar task using the select_if() function within the {dplyr} package and indicating that we want to select all columns without an NA.
frank %>% select_if(~!is.na(.x))
Build a shiny app that fluidly retains the columns without NA
Now that we have a few strategies for removing columns with NA, we can build a {shiny} app that allows the user to select a player and then the server fluidly will drop the columns with NA so that we don’t need to use a messy if/else chain.
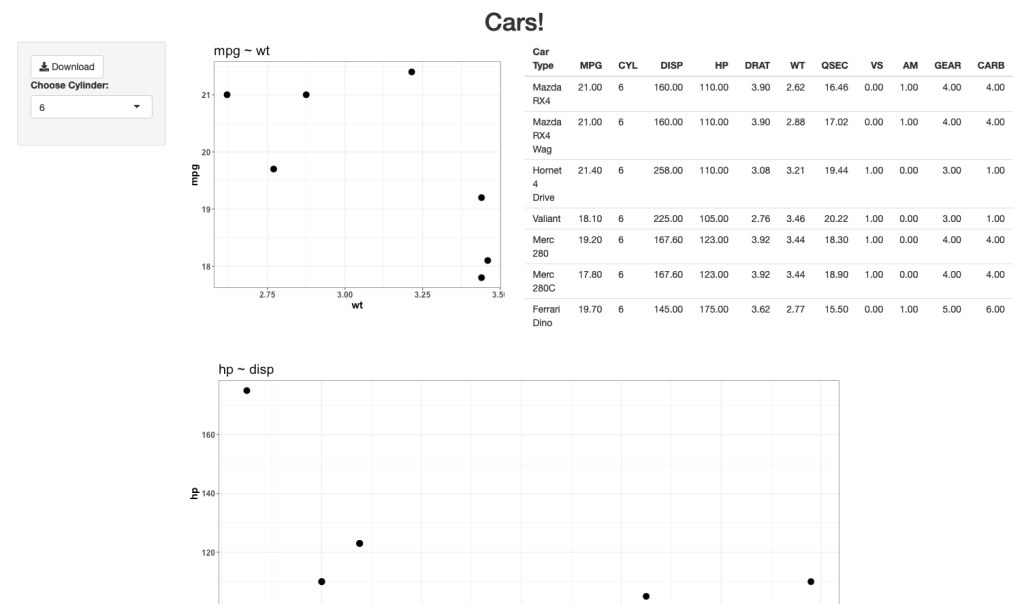
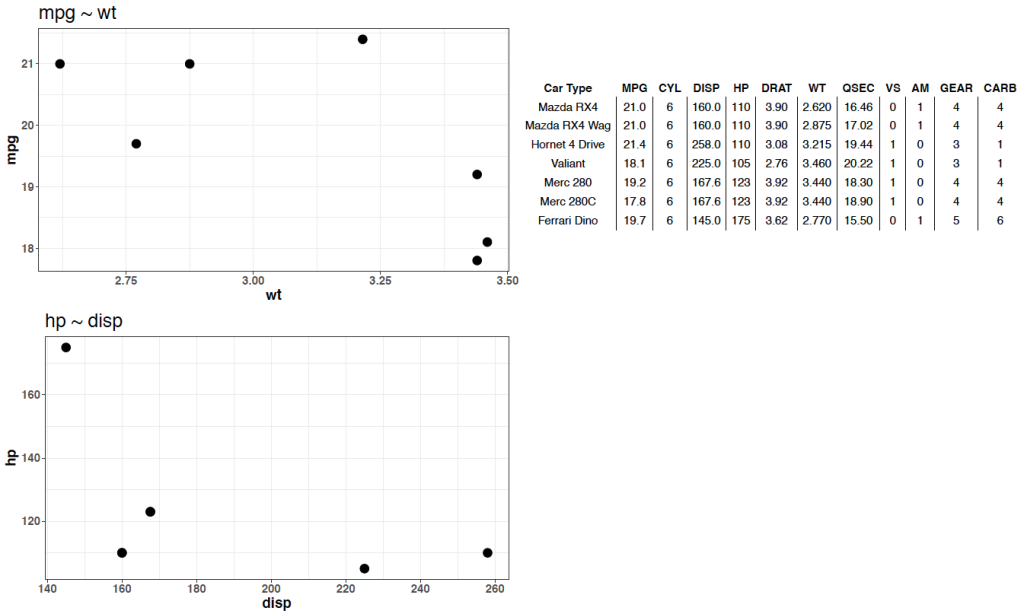
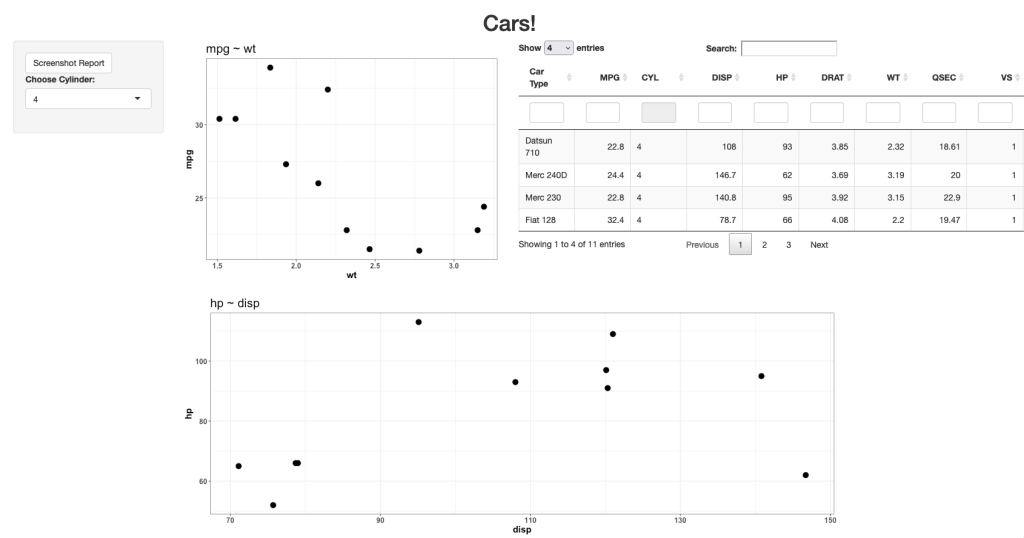
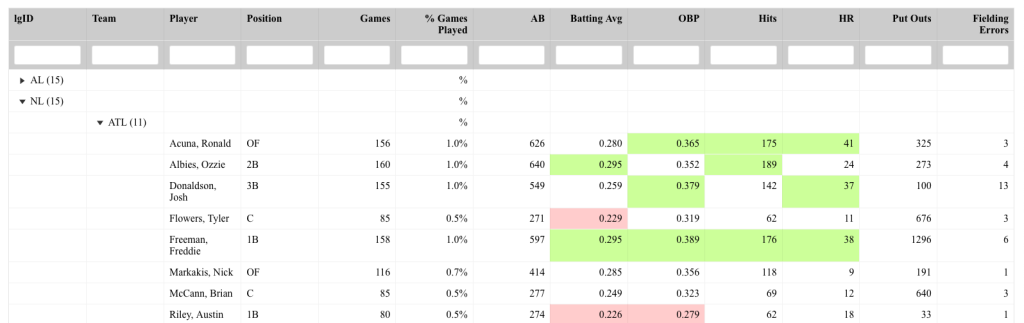
Notice that prior to dropping columns with NA I set the names of all of the columns in the table so that they look nicer when the table gets rendered. We can see from the figures that no matter which player is selected, the server intelligently drops columns with missing data, allowing the user to see only the statistics that are meaningful for the individual.
# UI
ui <- fluidPage(
sidebarPanel(
selectInput(inputId = 'player',
label = "Select Player",
choices = sort(unique(d$player)),
selected = FALSE,
multiple = FALSE)
),
mainPanel(
tableOutput(outputId = 'tbl')
)
)
# Server
server <- function(input, output){
# get selected player
dat_tbl <- reactive({ d %>%
filter(player == input$player)
})
# build table
output$tbl <- renderTable({ dat_tbl() %>%
setNames(c("Player", "Position", "Stat 1", "Stat 2", "Stat 3")) %>%
select_if(~!is.na(.x))
})
}
# deploy
shinyApp(ui, server)
Wrapping Up
Instead of having users see columns with NA, make your renderTable() function fluid and automatically drop columns with missing values to improve the user experience.
The full code is accessible on my GITHUB page.