Intro
Tim Newans and colleagues recently published a nice paper discussing the value of mixed models compared to repeated measures ANOVA in sport science research (1). I thought the paper highlighted some key points, though I do disagree slightly with the notion that sport science research hasn’t adopted mixed models, as I feel like they have been relatively common in the field over the past decade. That said, I wanted to do a blog that goes a bit further into mixed models because I felt like, while the aim of the paper was to show their value compared with repeated measures ANOVA, there are some interesting aspects of mixed models that weren’t touched upon in the manuscript. In addition to building up several mixed models, it might be fun to extend the final model to a Bayesian mixed model to show the parallels between the two and some of the additional things that we can learn with posterior distributions. The data used by Newans et al. had independent variables that were categorical, level of competition and playing position. The data I will use here is slightly different in that the independent variable is a continuous variable, but the concepts still apply.
Obtain the Data and Perform EDA
For this tutorial, I’ll use the convenient sleepstudy data set in the {lme4} package. This study is a series of repeated observations of reaction time on subjects that were being sleep deprived over 10 days. This sort of repeated nature of data is not uncommon in sport science as serial measurements (e.g., GPS, RPE, Wellness, HRV, etc) are frequently recorded in the applied environment. Additionally, many sport and exercise studies collect time point data following a specific intervention. For example, measurements of creatine-kinase or force plate jumps at intervals of 0, 24, and 48 hours following a damaging bout of exercise or competition.
knitr::opts_chunk$set(echo = TRUE)
library(tidyverse)
library(lme4)
library(broom)
library(gt)
library(patchwork)
library(arm)
theme_set(theme_bw())
dat <- sleepstudy dat %>% head()
The goal here is to obtain an estimate about the change in reaction time (longer reaction time means getting worse) following a number of days of sleep deprivation. Let’s get a plot of the average change in reaction time for each day of sleep deprivation.
dat %>%
group_by(Days) %>%
summarize(N = n(),
avg = mean(Reaction),
se = sd(Reaction) / sqrt(N)) %>%
ggplot(aes(x = Days, y = avg)) +
geom_ribbon(aes(ymin = avg - 1.96*se, ymax = avg + 1.96*se),
fill = "light grey",
alpha = 0.8) +
geom_line(size = 1.2) +
scale_x_continuous(labels = seq(from = 0, to = 9, by = 1),
breaks = seq(from = 0, to = 9, by = 1)) +
labs(x = "Days of Sleep Deprivation",
y = "Average Reaction Time",
title = "Reaction Time ~ Days of Sleep Deprivation",
subtitle = "Mean ± 95% CI")
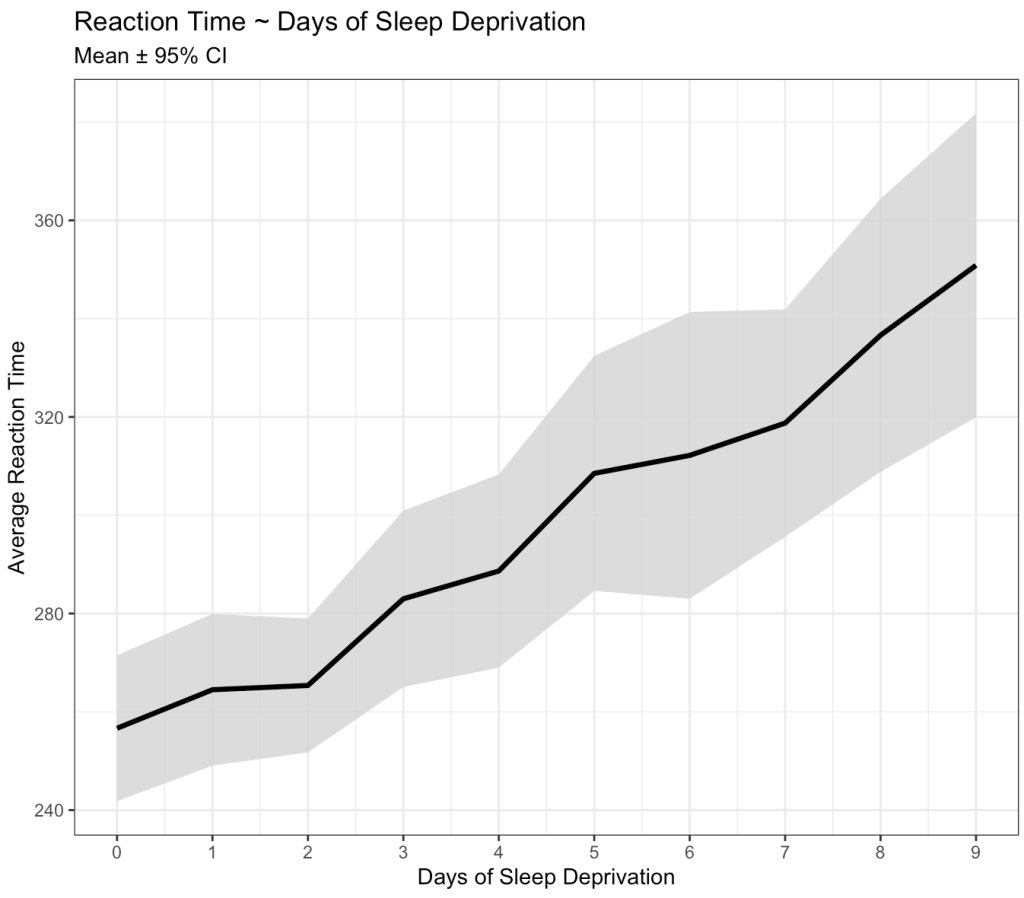
Okay, we can clearly see that something is going on here. As the days of sleep deprivation increase the reaction time in the test is also increasing, a fairly intuitive finding.
However, we also know that we are dealing with repeated measures on a number of subjects. As such, some subjects might have differences that vary from the group. For example, some might have a worse effect than the population average while others might not be that impacted at all. Let’s tease this out visually.
dat %>%
ggplot(aes(x = Days, y = Reaction)) +
geom_line(size = 1) +
geom_point(shape = 21,
size = 2,
color = "black",
fill = "white") +
geom_smooth(method = "lm",
se = FALSE) +
facet_wrap(~Subject) +
scale_x_continuous(labels = seq(from = 0, to = 9, by = 3),
breaks = seq(from = 0, to = 9, by = 3)) +
labs(x = "Days of Sleep Deprivation",
y = "Average Reaction Time",
title = "Reaction Time ~ Days of Sleep Deprivation")
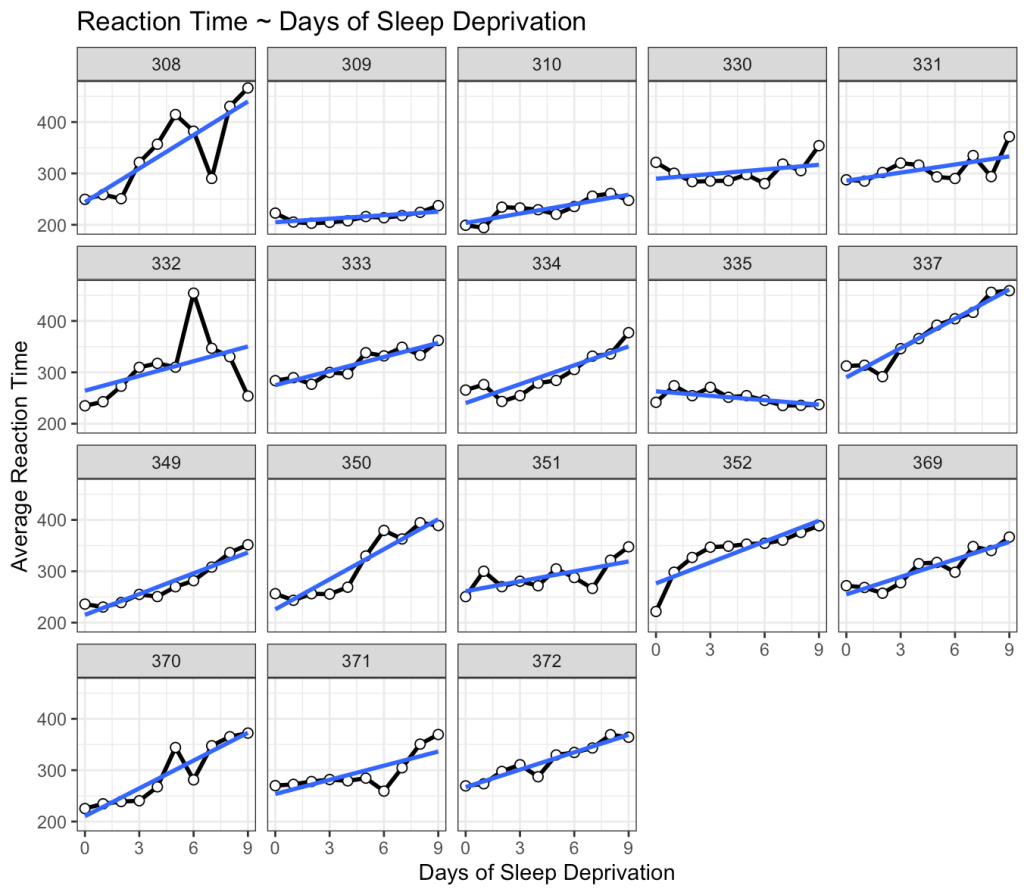
This is interesting. While we do see that many of the subjects exhibit an increase reaction time as the number of days of sleep deprivation increase there are a few subjects that behave differently from the norm. For example, Subject 309 seems to have no real change in reaction time while Subject 335 is showing a slight negative trend!
Linear Model
Before we get into modeling data specific to the individuals in the study, let’s just build a simple linear model. This is technically “the wrong” model because it violates assumptions of independence. After all, repeated measures on each individual means that there is going to be a level of correlation from one test to the next for any one individual. That said, building a simple model, even if it is the wrong model, is sometimes a good place to start just to help get your bearings, understand what is there, and have a benchmark to compare future models against. Additionally, the simple model is easier to interpret, so it is a good first step.
fit_ols <- lm(Reaction ~ Days, data = dat)
summary(fit_ols)
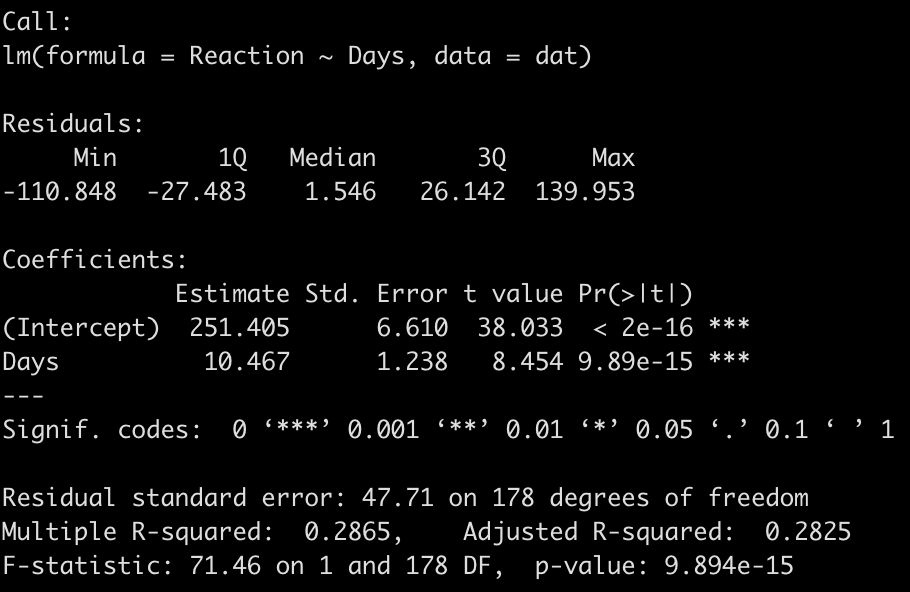
We end up with a model that suggests that for every one unit increase in a day of sleep deprivation, on average, we see about a 10.5 second increase in reaction time. The coefficient for the Days variable also has a standard error of 1.238 and the model r-squared indicates that days of sleep deprivation explain about 28.7% of the variance in reaction time.
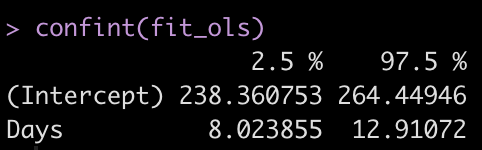
Let’s get the confidence intervals for the model intercept and Days coefficient.
confint(fit_ols)
Summary Measures Approach
Before moving to the mixed model approach, I want to touch on simple approach that was written about in 1990 by Matthews and Altman (2). I was introduced to this approach by one of my former PhD supervisors, Matt Weston, as he used it to analyze data in 2011 for a paper with my former lead PhD supervisor, Barry Drust, where they were quantifying the intensity of Premier League match-play for players and referees. This approach is extremely simple and, while you may be asking yourself, “Why not just jump into the mixed model and get on with it?”, just bear with me for a moment because this will make sense later when we begin discussing pooling effects in mixed models and Bayesian mixed models.
The basic approach suggested by Matthews (2) for this type of serially collected data is to treat each individual as the unit of measure and identify a single number, which summarizes that subject’s response over time. For our analysis here, the variable that we are interested in for each subject is the Days slope coefficient, as this value tells us the rate of change in reaction time for every passing day of sleep deprivation. Let’s construct a linear regression for each individual subject in the study and place their slope coefficients into a table.
ind_reg <- dat %>%
group_by(Subject) %>%
group_modify(~ tidy(lm(Reaction ~ Days, data = .x)))
ind_days_slope <- ind_reg %>%
filter(term == "Days")
ind_days_slope %>%
ungroup() %>%
gt() %>%
fmt_number(columns = estimate:statistic,
decimals = 2) %>%
fmt_number(columns = p.value,
decimals = 3)
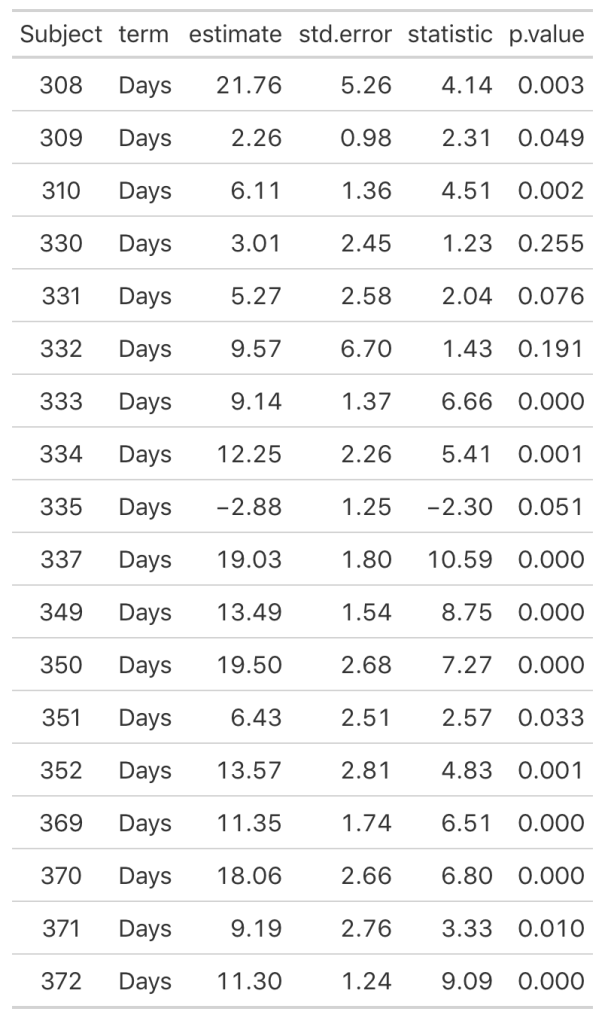
As we noticed in our visual inspection, Subject 335 had a negative trend and we can confirm their negative slope coefficient (-2.88). Subject 309 had a relatively flat relationship between days of sleep deprivation and reaction time. Here we see that their coefficient is 2.26 however the standard error, 0.98, indicates rather large uncertainty [95% CI: 0.3 to 4.22].
Now we can look at how each Subject’s response compares to the population. If we take the average of all of the slopes we get basically the same value that we got from our OLS model above, 10.47. I’ll build two figures, one that shows each Subject’s difference from the population average of 10.47 and one that shows each Subject’s difference from the population centered at 0 (being no difference from the population average). Both tell the same story, but offer different ways of visualizing the subjects relative to the population.
pop_avg
plt_to_avg <- ind_days_slope %>%
mutate(pop_avg = pop_avg,
diff = estimate - pop_avg) %>%
ggplot(aes(x = estimate, y = as.factor(Subject))) +
geom_vline(xintercept = pop_avg) +
geom_segment(aes(x = diff + pop_avg,
xend = pop_avg,
y = Subject,
yend = Subject),
size = 1.2) +
geom_point(size = 4) +
labs(x = NULL,
y = "Subject",
title = "Difference compared to population\naverage change in reaction time (10.47 sec)")
plt_to_zero <- ind_days_slope %>%
mutate(pop_avg = pop_avg,
diff = estimate - pop_avg) %>%
ggplot(aes(x = diff, y = as.factor(Subject))) +
geom_vline(xintercept = 0) +
geom_segment(aes(x = 0,
xend = diff,
y = Subject,
yend = Subject),
size = 1.2) +
geom_point(size = 4) +
labs(x = NULL,
y = "Subject",
title = "Difference compared to population\naverage reflected against a 0 change")
plt_to_avg | plt_to_zero
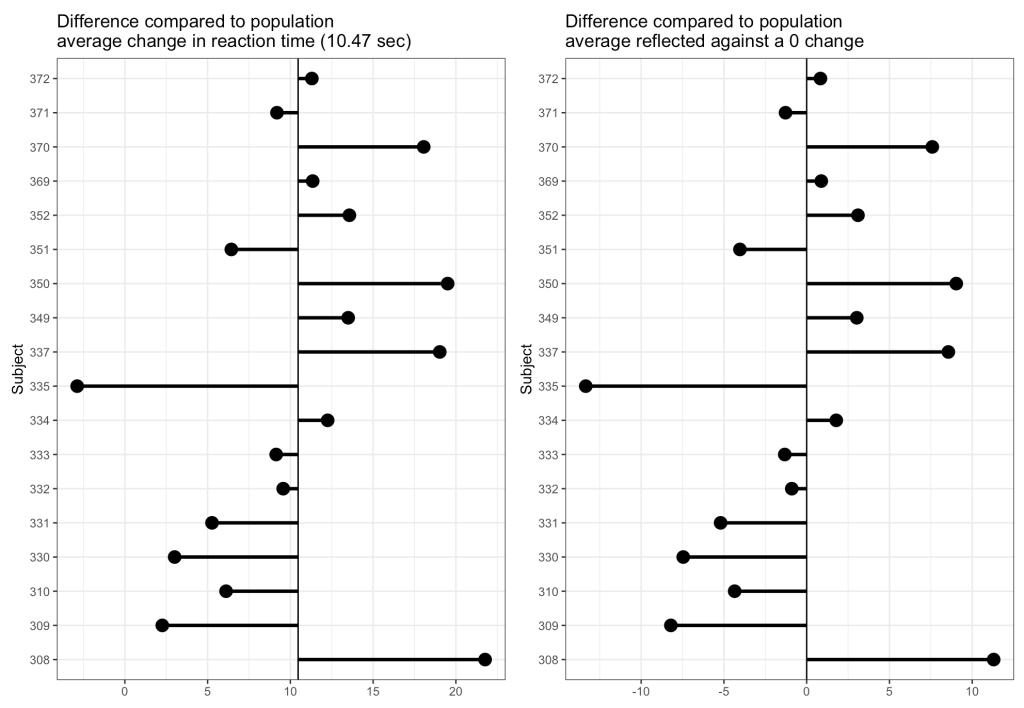
From the plots, it is clear to see how each individual behaves compared to the population average. Now let’s build some mixed models and compare the results.
Mixed Models
The aim of the mixed model here is to in someway acknowledge that we have these repeated measures on each individual and we need to account for that. In the simple linear model approach above, the Subjects shared the same variance, which isn’t accurate given that each subject behaves slightly differently, which we saw in our initial plot and in the individual linear regression models we constructed in the prior section. Our goal is to build a model that allows us to make an inference about the way in which the amount of sleep deprivation, measured in days, impacts a human’s reaction time performance. Therefore, we wouldn’t want to add each subject into a single regression model, creating a coefficient for each individual within the model, as that would be akin to making comparisons between each individual similar to what we would do in an ANOVA (and it will also use a lot of degrees of freedom). So, we want to acknowledge that there are individual subjects that may vary from the population, while also modeling our question of interest (Reaction Time ~ Days of Sleep Deprivation).
Intercept Only Model
We begin with a simple model that has an intercept only but allows that intercept to vary by subject. As such, this model is not accounting for days of sleep deprivation. Rather, it is simply telling us the average reaction time response for the group and how each subject varies from that response.
fit1 <- lmer(Reaction ~ 1 + (1|Subject), data = dat)
display(fit1)
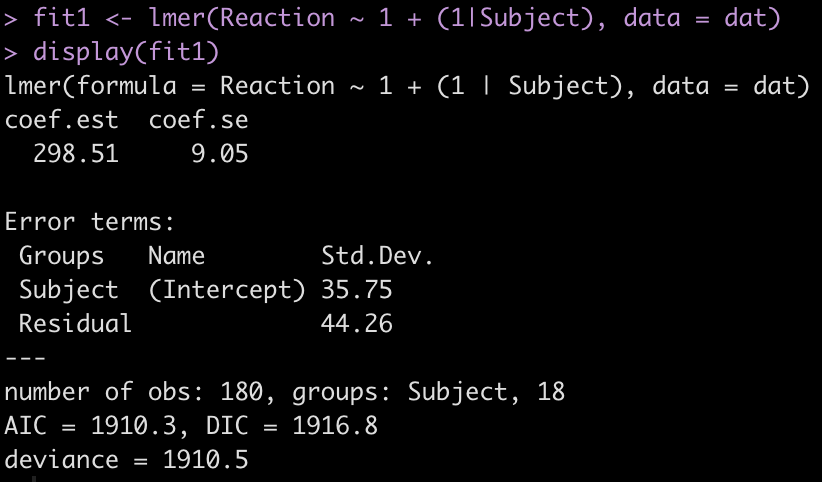
The output provides us with an average estimate for reaction time of 298.51. Again, this is simply the group average for reaction time. We can confirm this easily.
mean(dat$Reaction)
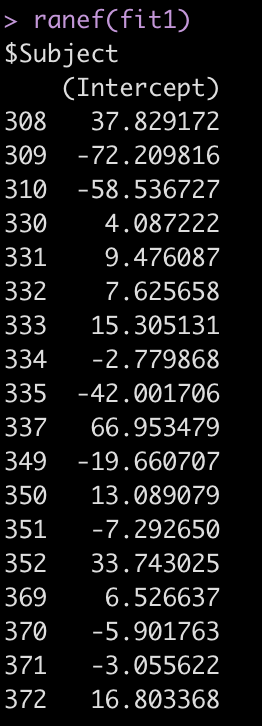
The Error terms in our output above provide us with the standard deviation around the population intercept, 35.75, which is the between-subject variability. The residual here represents our within-subject standard variability. We can extract the actual values for each subject. Using the ranef() function we get the difference between each subject and the population average intercept (the fixed effect intercept).
ranef(fit1)
One Fixed Effect plus Random Intercept Model
We will now extend the above mixed model to reflect our simple linear regression model except with random effects specified for each subject. Again, the only thing we are allowing to vary here is the individual subject’s intercept for reaction time.
fit2 <- lmer(Reaction ~ Days + (1|Subject), data = dat)
display(fit2)
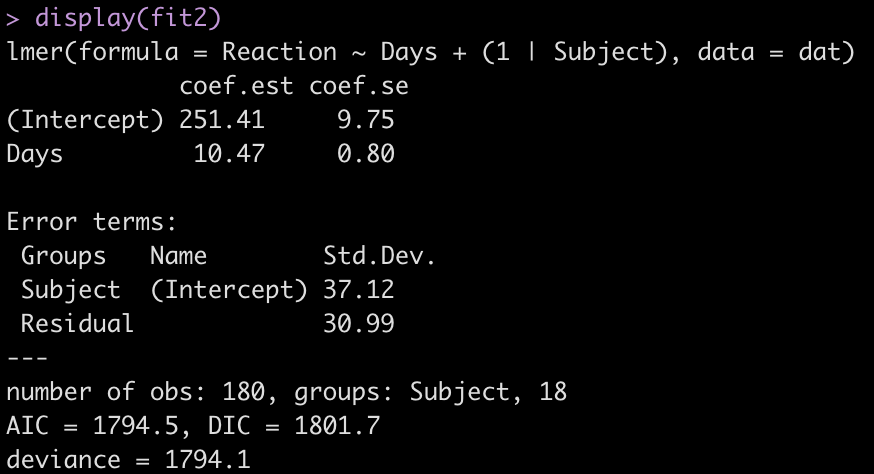
Adding the independent variable, Days, has changed the intercept, decreasing it from 298.51 to 251.41. The random effects have also changed from the first model. In the intercept only model we had an intercept standard deviation of 35.75 with a residual standard deviation of 44.26. In this model, accounting for days of sleep deprivation, the random effects intercept is now 37.12 and the residual (within subject) standard deviation has dropped to 30.99. The decline in the residual standard deviation tells us the model is picking up some of the individual differences that we have observed in plot of each subjects’ data.
Let’s look at the random effects intercept for each individual relative to the fixed effect intercept.
ranef(fit2)
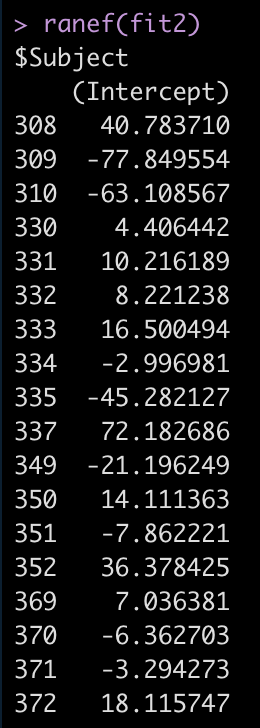
For example, Subject 309 has an intercept that is 77.8 seconds below the fixed effect intercept, while Subject 308 is 40.8 seconds above the fixed effect intercept.
If we use the coef() function we get returned the actual individual linear regression equation for each subject. Their difference compared to the population will be added to the fixed effect intercept, to create their individual intercept, and we will see the days coefficient, which is the same for each subject because we haven’t specified a varying slope model (yet).
coef(fit2)
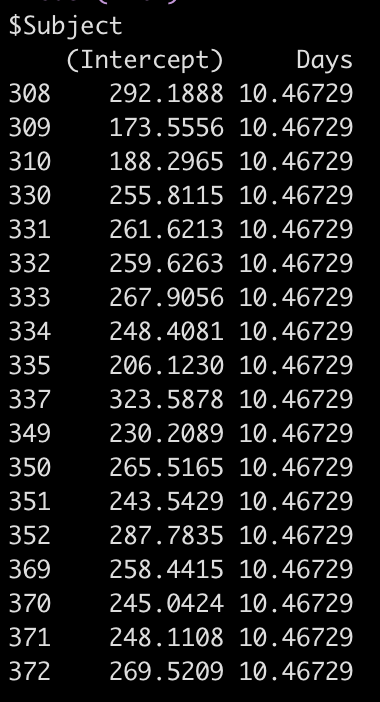
So, for example, the equation for Subject 308 has an equation of:
292.19 + 10.47*Days
Let’s also compare the results of this model to our original simple regression model. I’ll put all of the model results so far into a single data frame that we can keep adding to as we go, allowing us to compare the results.
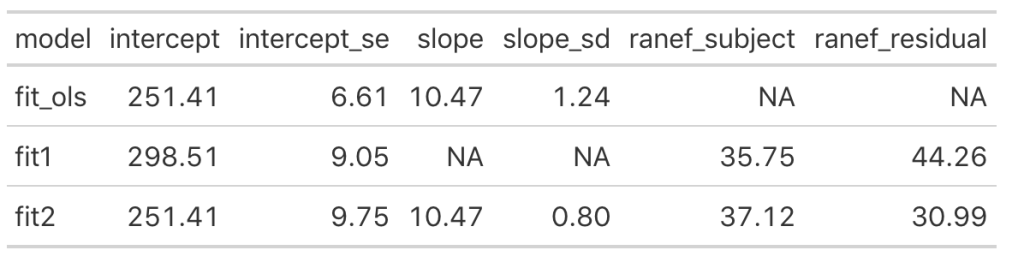
We can see that once we add the day variable to the model (in fit_ols, and fit2) the model intercept for reaction time changes to reflect its output being dependent on this information. Notice that the intercept and slope values between the simple linear regression model and the fit2 model are the same. The mean result hasn’t changed but notice that the standard error for the intercept and slope as been altered. In particular, notice how much the standard error of the slope variable has declined once we moved from a simple regression to a mixed model. This is due do the fact that the model now knows that there are individuals with repeated measurements being recorded.
Looking at the random effects changes between fit1 (intercept only model) and fit2, we see that we’ve increased in standard deviation for the intercept (35.75 to 37.12), which represents the between-subject variability. Additionally, we have substantially decreased the residual standard deviation (44.26 to 30.99), which represents our within-subject variability. Again, because we have not explicitly told the model that certain measurements are coming from certain individuals, there appears to be more variability between subjects and less variability within-subject. Basically, there is some level of autocorrelation to the individual’s reaction time performance whereby they are more similar to themselves than they are to other people. This makes intuitive sense — if you can’t agree with yourself, who can you agree with?!
Random Slope and Intercept Model
The final mixed model we will build will allow not only the intercept to vary randomly between subjects but also the slope. Recall above that the coefficient for Days was the same across all subjects. This is because that model allowed the subjects to have different model intercepts but it made the assumption that they had the same model slope. However, we may have reason to believe that the slopes also differ between subjects after looking at the individual subject plots in section 1.
fit3 <- lmer(Reaction ~ Days + (1 + Days|Subject), data = dat)
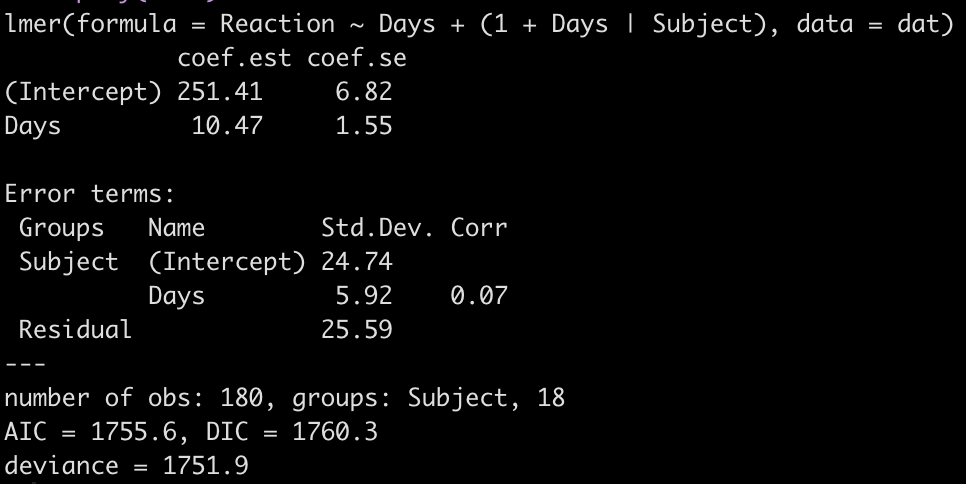
display(fit3)
Let’s take a look at the regression model for each individual.
coef(fit3)
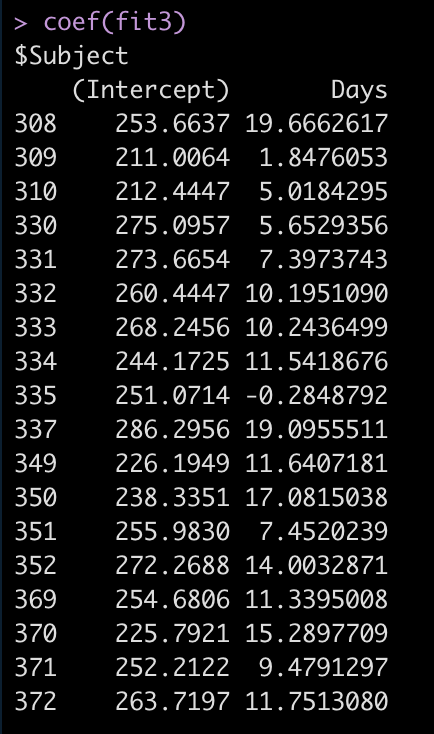
Now we see that the coefficient for the Days variable is different for each Subject. Let’s add the results of this model to our results data frame and compare everything.
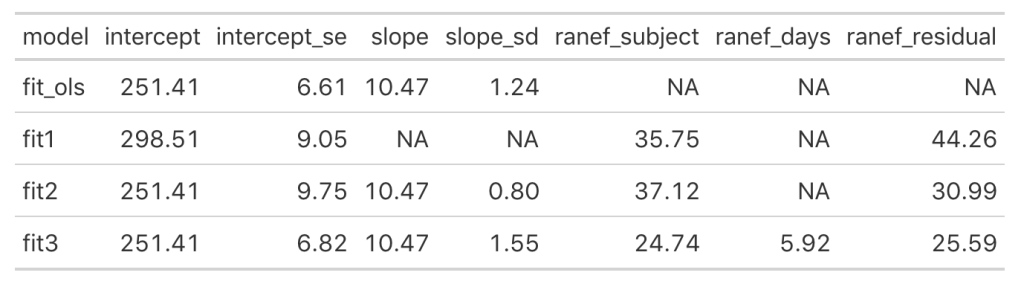
Looking at this latest model we see that the intercept and slope coefficients have remained unchanged relative to the other models. Again, the only difference in fixed effects comes at the standard error for the intercept and the slope. This is because the variance in the data is being partitioned between the population estimate fixed effects and the individual random effects. Notice that for the random effects in this model, fit3, we see a substantial decline in the between-subject intercept, down to 24.74 from the mid 30’s in the previous two models. We also see a substantial decline the random effect residual, because we are now seeing less within individual variability as we account for the random slope. The addition here is the random effect standard deviation for the slope coefficient, which is 5.92.
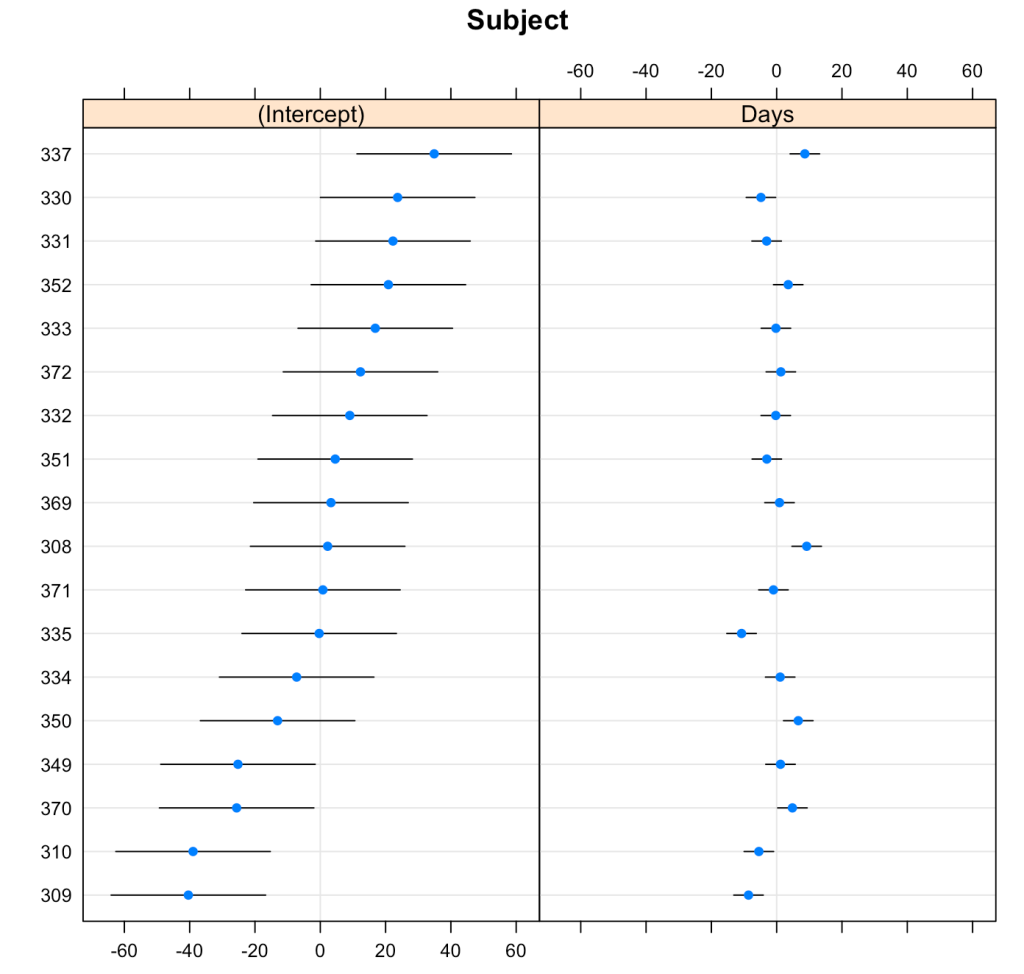
We can plot the results of the varying intercepts and slopes across subjects using the {lattice} package.
lattice::dotplot(ranef(fit3, condVar = T))
Comparing the models
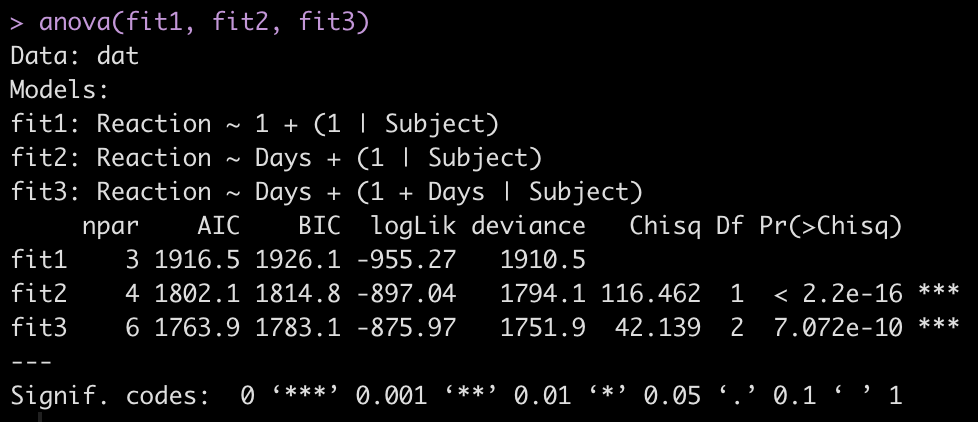
To make model comparisons, Newans and colleagues used Akaike Information Criterion (AIC) to determine which model fit the data best. With the anova() function we simply pass our three mixed models in as arguments and we get returned several model comparison metrics including AIC, BIC, and p-values. These values are lowest for fit3, indicating it is the better model at explaining the underlying data. Which shouldn’t be too surprising given how individual the responses looked in the initial plot of the data.
anova(fit1, fit2, fit3)
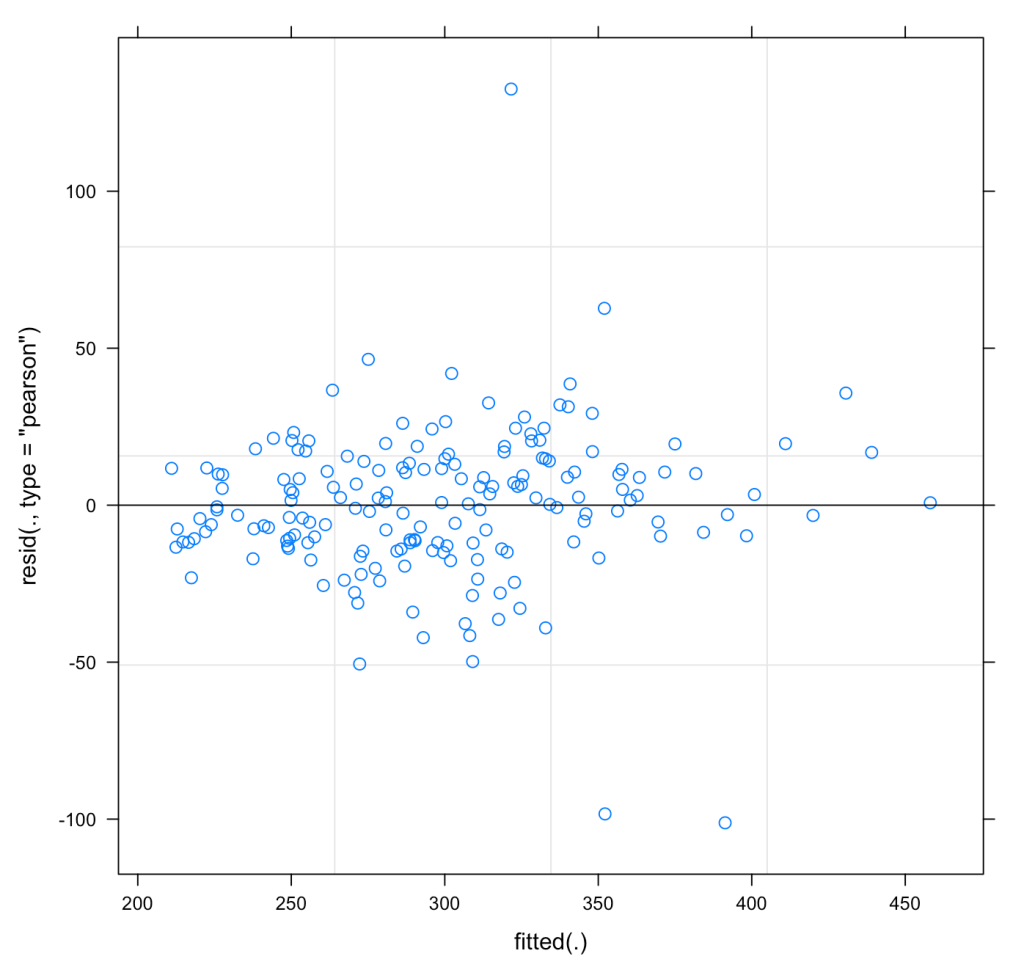
We can also plot the residuals of fit3 to see whether they violate any assumptions of homoscedasticity or normality.
plot(fit3)
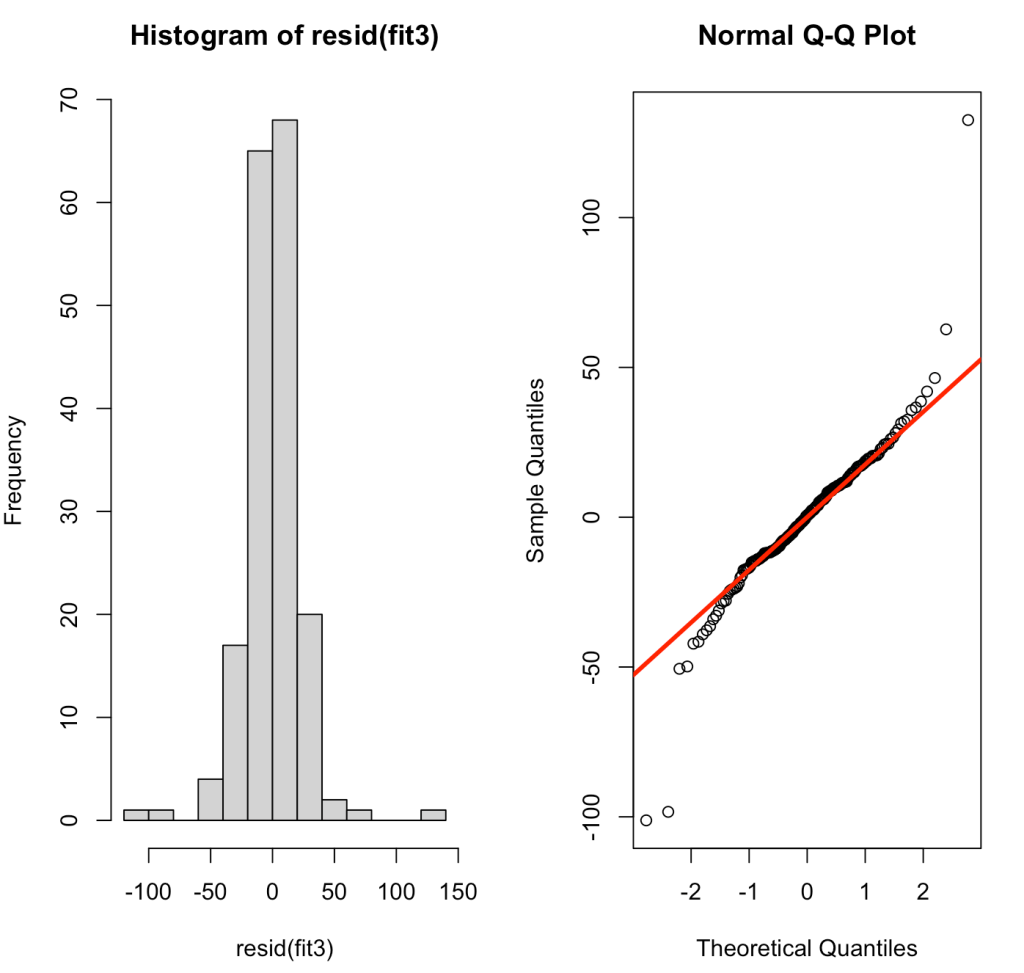
par(mfrow = c(1,2))
hist(resid(fit3))
qqnorm(resid(fit3))
qqline(resid(fit3), col = "red", lwd = 3)
Pooling Effects
So what’s going on here? The mixed model is really helping us account for the repeated measures and the correlated nature of an individuals data. In doing so, it allows us to make an estimate of a population effect (fixed effect), while adjusting the standard errors to be more reasonable given the violation of independence assumption. The random effects allow us to see how much variance there is for each individual measured within the population. In a way, I tend to think about mixed models as a sort of bridge to Bayesian analysis. In my mind, the fixed effects seem to look a bit like priors and the random effects are the adjustment we make to those priors having seen some data for an individual. In our example here, each of the 18 subjects have 10 reaction time measurements. If we were dealing with a data set that had different numbers of observations for each individual, however, we would see that those with less observations are pulled closer towards the population average (the fixed effect coefficients) while those with a larger number of observations are allowed to deviate further from the population average, if they indeed are different. This is because with more observations we have more confidence about what we are observing for that individual. In effect, this is called pooling.
In their book, Data Analysis Using Regression and Multilevel/Hierarchical Models, Gelman and Hill (4) discuss no-, complete-, and partial-pooling models.
- No-pooling models occur when the data from each subject are analyzed separately. This model ignores information in the data and could lead to poor inference.
- Complete-pooling disregards any variation that might occur between subjects. Such suppression of between-subject variance is missing the point of conducting the analysis and looking at individual difference.
- Partial-pooling strikes a balance between the two, allowing for some pooling of population data (fixed effects) while honoring the fact that there are individual variations occurring within the repeated measurements. This type of analysis is what we obtain with a mixed model.
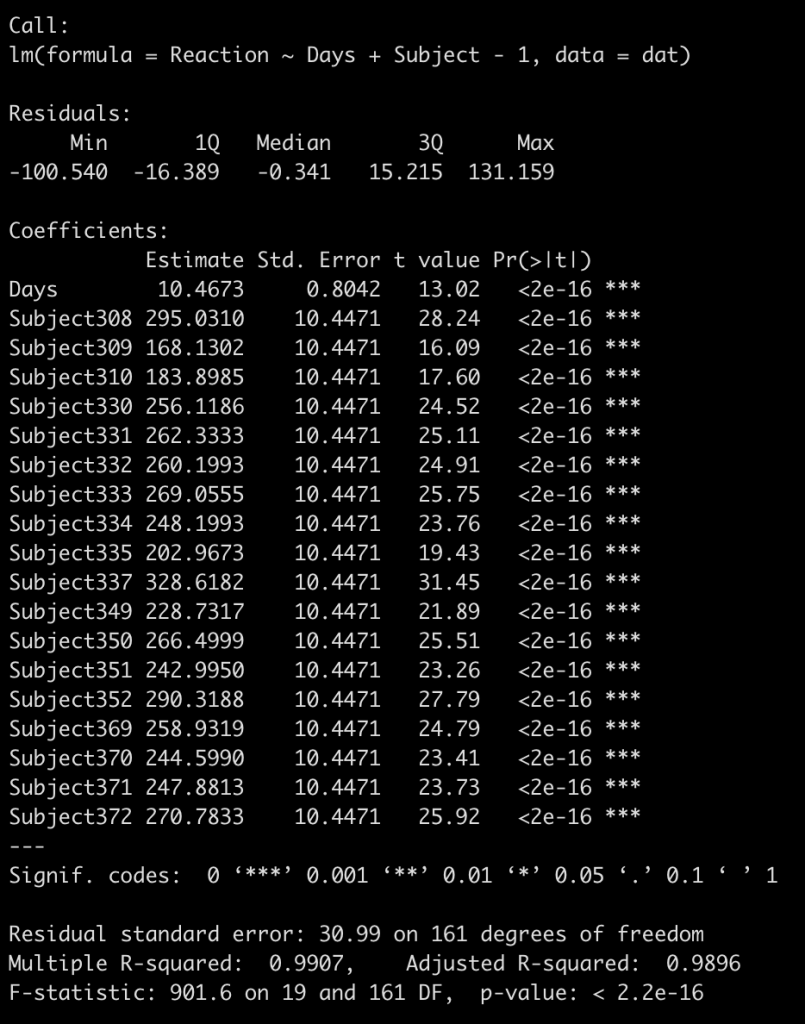
I find it easiest to understand this by visualizing it. We already have a complete-pooling model (fit_ols) and a partial-pooling model (fit3). We need to fit the no-pooling model, which is a regression equation with each subject entered as a fixed effect. As a technical note, I will also add to the model -1 to have a coefficient for each subject returned instead of a model intercept.
fit_no_pool <- lm(Reaction ~ Days + Subject - 1, data = dat)
summary(fit_no_pool)
To keep the visual small, we will fit each of our three models to 4 subjects (308, 309, 335, and 331) and then plot the respective regression lines. In addition, I will also plot the regression line from the individualized regression/summary measures approach that we built first, just to show the difference.
## build a data frame for predictions to be stored
Subject <- as.factor(c(308, 309, 335, 331))
Days <- seq(from = 0, to = 9, by = 1)
pred_df <- crossing(Subject, Days)
pred_df
## complete pooling predictions
pred_df$complete_pool_pred <- predict(fit_ols, newdata = pred_df)
## no pooling predictions
pred_df$no_pool_pred <- predict(fit_no_pool, newdata = pred_df)
## summary measures/individual regression
subject_coefs <- ind_reg %>%
filter(Subject %in% unique(pred_df$Subject)) %>%
dplyr::select(Subject, term, estimate) %>%
mutate(term = ifelse(term == "(Intercept)", "Intercept", "Days")) %>%
pivot_wider(names_from = term,
values_from = estimate)
subject_coefs %>%
as.data.frame()
pred_df <- pred_df %>%
mutate(ind_reg_pred = ifelse(
Subject == 308, 244.19 + 21.8*Days,
ifelse(
Subject == 309, 205.05 + 2.26*Days,
ifelse(
Subject == 331, 285.74 + 5.27*Days,
ifelse(Subject == 335, 263.03 - 2.88*Days, NA)
)
)
))
## partial pooling predictions
pred_df$partial_pool_pred <- predict(fit3,
newdata = pred_df,
re.form = ~(1 + Days|Subject))
## get original results and add to the predicted data frame
subject_obs_data <- dat %>%
filter(Subject %in% unique(pred_df$Subject)) %>%
dplyr::select(Subject, Days, Reaction)
pred_df <- pred_df %>%
left_join(subject_obs_data)
## final predicted data set with original observations
pred_df %>%
head()
## plot results
pred_df %>%
pivot_longer(cols = complete_pool_pred:partial_pool_pred,
names_to = "model_pred") %>%
arrange(model_pred) %>%
ggplot(aes(x = Days, y = Reaction)) +
geom_point(size = 4,
shape = 21,
color = "black",
fill = "white") +
geom_line(aes(y = value, color = model_pred),
size = 1.1) +
facet_wrap(~Subject)
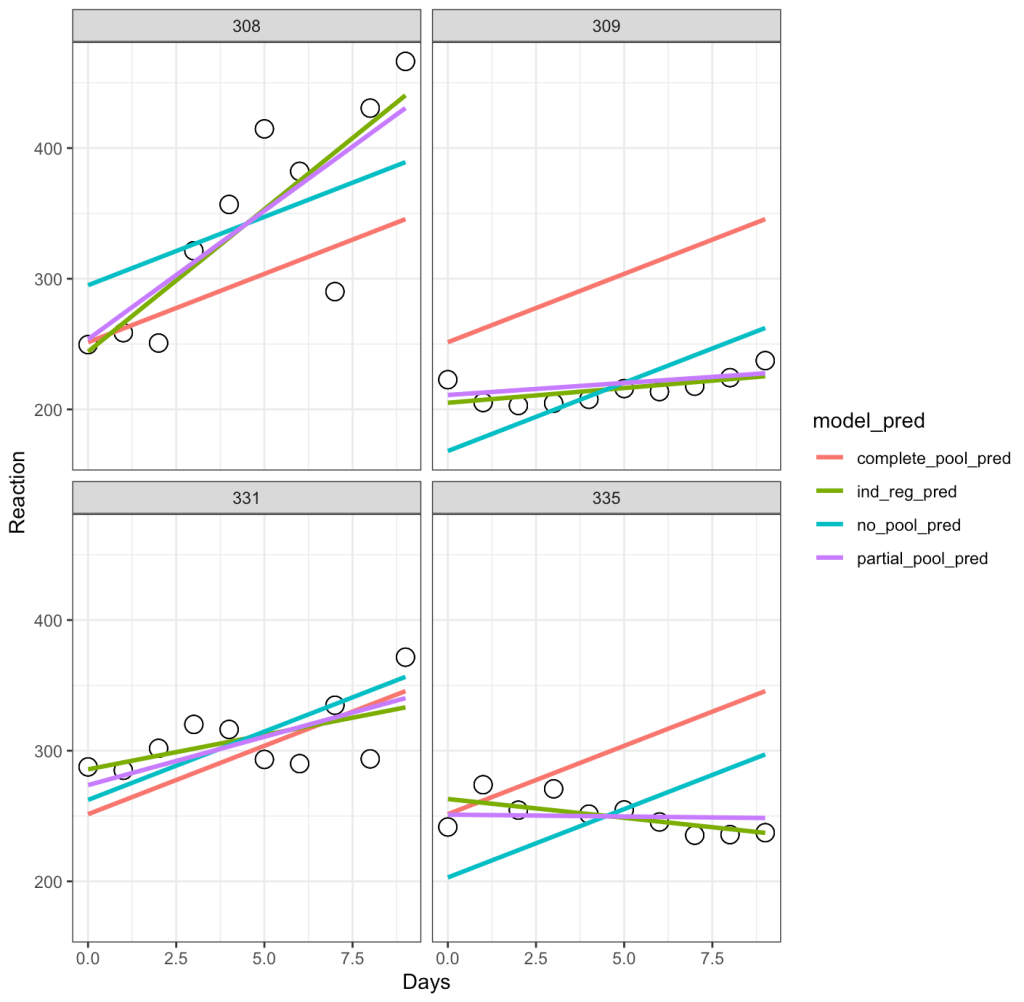
Let’s unpack this a bit:
- The complete-pooling line (orange) has the same intercept and slope for each of the subjects. Clearly it does not fit the data well for each subject.
- The no-pooling line (dark green) has the same slope for each subject but we notice that the intercept varies as it is higher for some subject than others.
- The partial-pooling line (purple) comes from the mixed model and we can see that it has a slope and intercept that is unique to each subject.
- Finally, the independent regression/summary measures line (light green) is similar to the partial-pooling line, as a bespoke regression model was built for each subject. Note, that in this example the two lines have very little difference but, if we were dealing with subjects who had varying levels of sample size, this would not be the case. The reason is because those with lower sample size will have an independent regression line completely estimated from their observed data, however, their partial pooling line will be pulled more towards the population average, given their lower sample.
Bayesian Mixed Models
Since I mentioned that I tend to think of mixed models as sort of a bridge to the Bayesian universe, let’s go ahead at turn our mixed model into a Bayes model and see what else we can do with it.
I’ll keep things simple here and allow the {rstanarm} library to use the default, weakly informative priors.
library(rstanarm)
fit_bayes <- stan_lmer(Reaction ~ Days + (1 + Days|Subject), data = dat)
fit_bayes
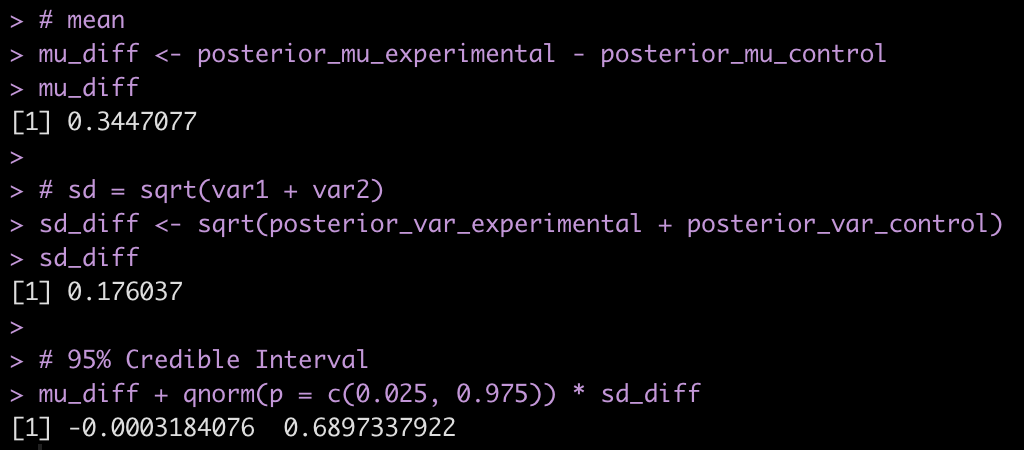
# If you want to extract the mean, sd and 95% Credible Intervals
# summary(fit_bayes,
# probs = c(0.025, 0.975),
# digits = 2)
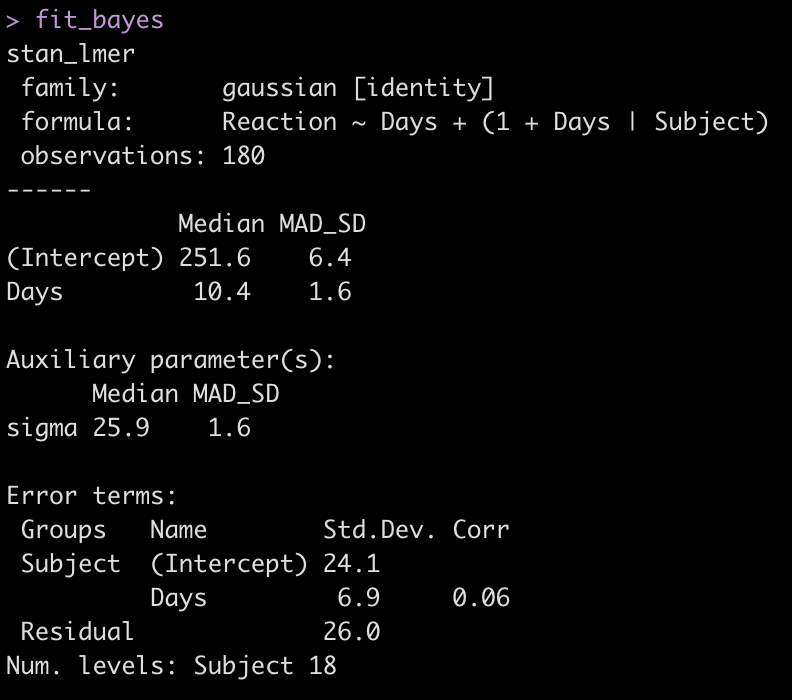
Let’s add the model output to our comparison table.
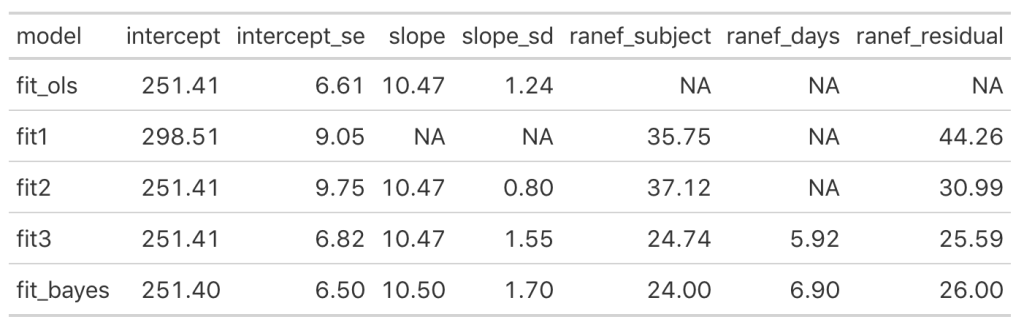
We see some slight changes between fit3 and fit_bayes, but nothing that drastic.
Let’s see what the posterior draws for all of the parameters in our model look like.
# Extract the posterior draws for all parameters
post_sims <- as.matrix(fit_bayes)
dim(post_sims)
head(post_sims)
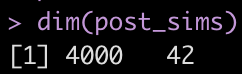
Yikes! That is a large matrix of numbers (4000 rows x 42 columns). Each subject gets, by default, 4000 draws from the posterior and each column represents a posterior sample from each of the different parameters in the model.
What if we slim this down to see what is going on? Let’s see what possible parameters in our matrix are in our matrix we might be interested in extracting.
colnames(post_sims)
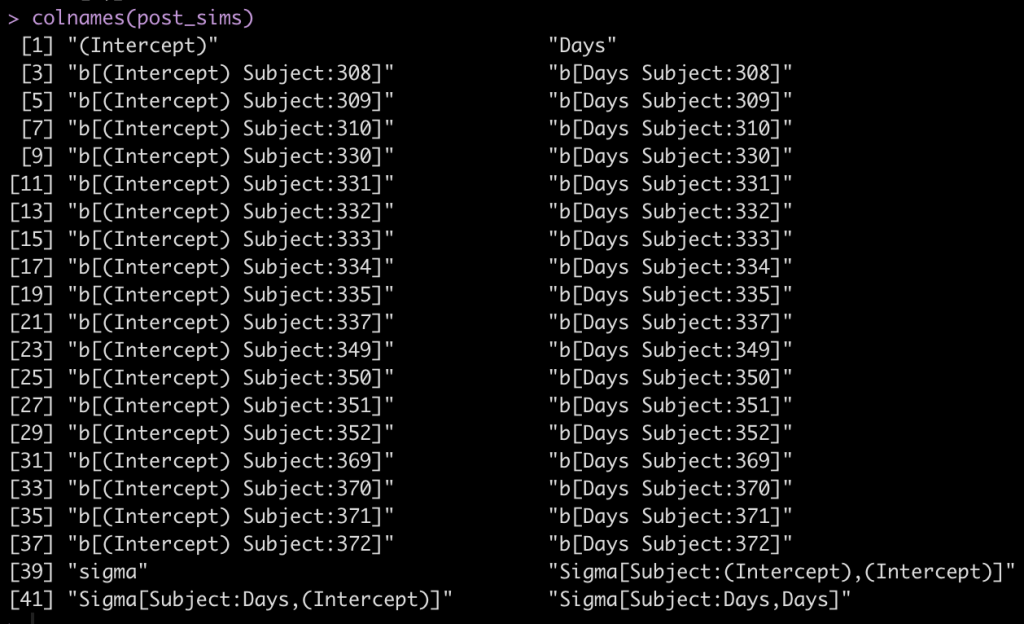
The first two columns are the fixed effect intercept and slope. Following that, we see that each subject has an intercept value and a Days value, coming from the random effects. Finally, we see that column names 39 to 42 are specific to the sigma values of the model.
Let’s keep things simple and see what we can do with the individual subject intercepts. Basically, we want to extract the posterior distribution of the fixed effects intercept and the posterior distribution of the random effects intercept per subject and combine those to reflect the posterior distribution of reaction time by subject.
## posterior draw from the fixed effects intercept
fixed_intercept_sims <- as.matrix(fit_bayes,
pars = "(Intercept)")
## posterior draws from the individual subject intercepts
subject_ranef_intercept_sims <- as.matrix(fit_bayes,
regex_pars = "b\\[\\(Intercept\\) Subject\\:")
## combine the posterior draws of the fixed effects intercept with each individual
posterior_intercept_sims <- as.numeric(fixed_intercept_sims) + subject_ranef_intercept_sims
head(posterior_intercept_sims[, 1:4])
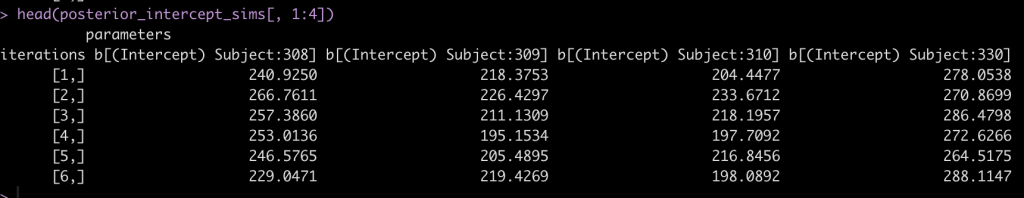
After drawing our posterior intercepts, we can get the mean, standard deviation, median, and 95% Credible Interval of the 4000 simulations for each subject and then graph them in a caterpillar plot.
## mean per subject
intercept_mean <- colMeans(posterior_intercept_sims)
## sd per subject
intercept_sd <- apply(X = posterior_intercept_sims,
MARGIN = 2,
FUN = sd)
## median and 95% credible interval per subject
intercept_ci <- apply(X = posterior_intercept_sims,
MARGIN = 2,
FUN = quantile,
probs = c(0.025, 0.50, 0.975))
## summary results in a single data frame
intercept_ci_df <- data.frame(t(intercept_ci))
names(intercept_ci_df) <- c("x2.5", "x50", "x97.5")
## Combine summary statistics of posterior simulation draws
bayes_df <- data.frame(intercept_mean, intercept_sd, intercept_ci_df)
round(head(bayes_df), 2)
## Create a column for each subject's ID
bayes_df$subject <- rownames(bayes_df)
bayes_df$subject <- extract_numeric(bayes_df$subject)
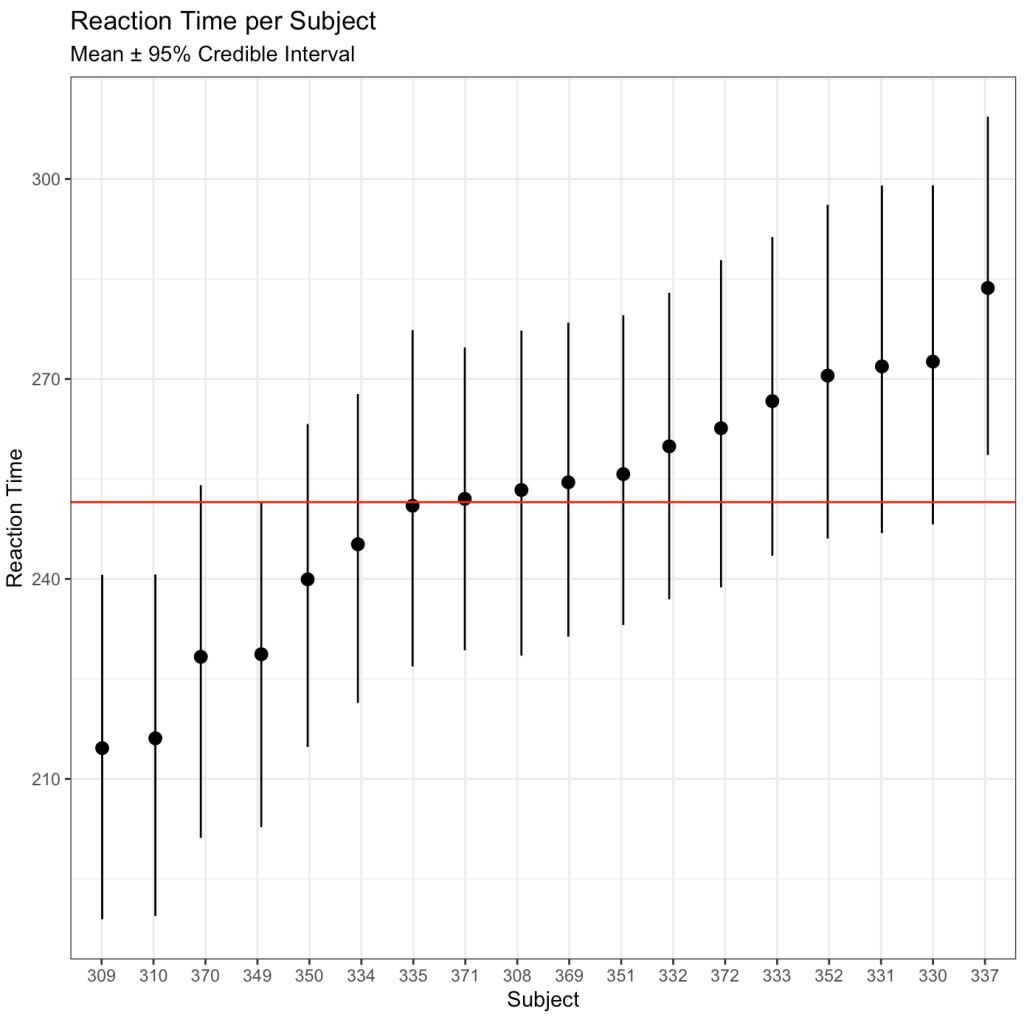
## Catepillar plot
ggplot(data = bayes_df,
aes(x = reorder(as.factor(subject), intercept_mean),
y = intercept_mean)) +
geom_pointrange(aes(ymin = x2.5,
ymax = x97.5),
position = position_jitter(width = 0.1,
height = 0)) +
geom_hline(yintercept = mean(bayes_df$intercept_mean),
size = 0.5,
col = "red") +
labs(x = "Subject",
y = "Reaction Time",
title = "Reaction Time per Subject",
subtitle = "Mean ± 95% Credible Interval")
We can also use the posterior distributions to compare two individuals. For example, let’s compare Subject 308 (column 1 of our posterior sim matrix) to Subject 309 (column 2 of our posterior sim matrix).
## create a difference between distributions
compare_dist <- posterior_intercept_sims[, 1] - posterior_intercept_sims[, 2]
# summary statistics of the difference
mean_diff <- mean(compare_dist)
sd_diff <- sd(compare_dist)
quantile_diff <- quantile(compare_dist, probs = c(0.025, 0.50, 0.975))
quantile_diff <- data.frame(t(quantile_diff))
names(quantile_diff) <- c("x2.5", "x50", "x97.5")
diff_df <- data.frame(mean_diff, sd_diff, quantile_diff)
round(diff_df, 2)
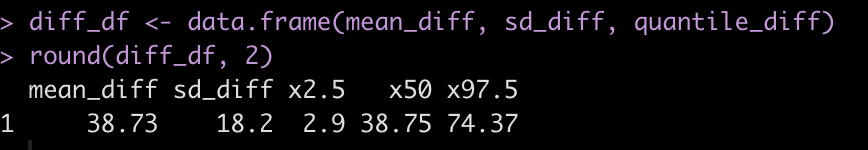
Subject 308 exhibits a 38.7 second higher reaction time than Subject 309 [95% Credible Interval 2.9 to 74.4].
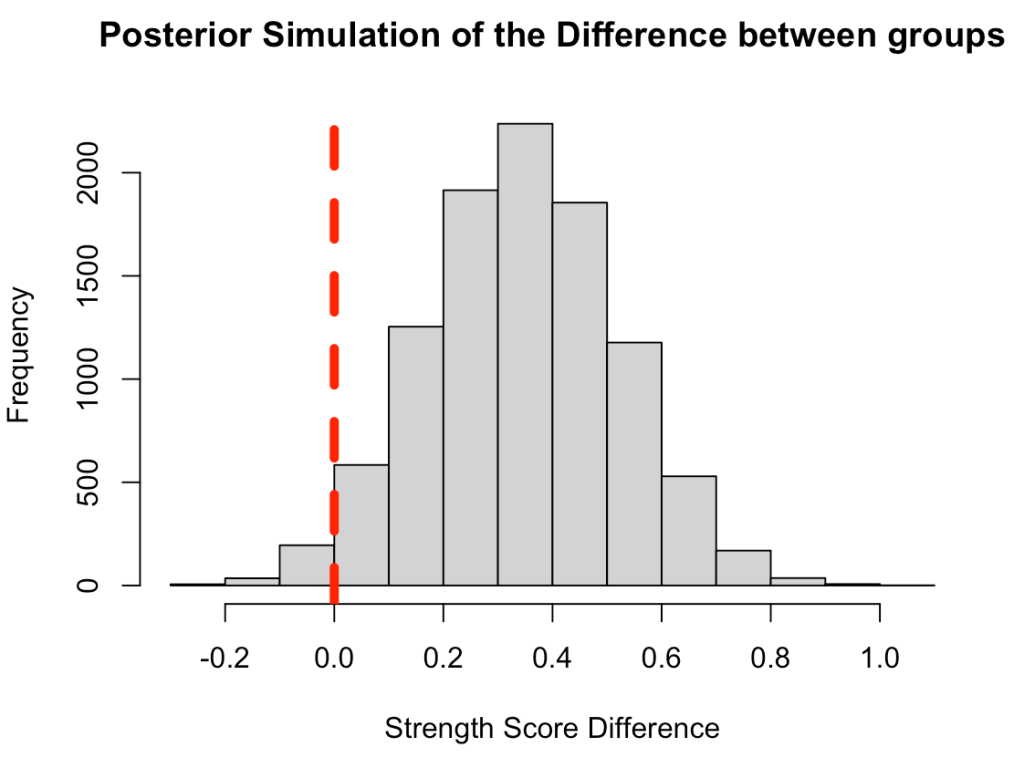
We can plot the posterior differences as well.
# Histogram of the differences
hist(compare_dist,
main = "Posterior Distribution Comparison\n(Subject 308 - Subject 309)",
xlab = "Difference in 4000 posterior simulations")
abline(v = 0,
col = "red",
lty = 2,
lwd = 2)
abline(v = mean_diff,
col = "black",
lwd = 2)
abline(v = quantile_diff$x2.5,
col = "black",
lty = 2,
lwd = 2)
abline(v = quantile_diff$x97.5,
col = "black",
lty = 2,
lwd = 2)
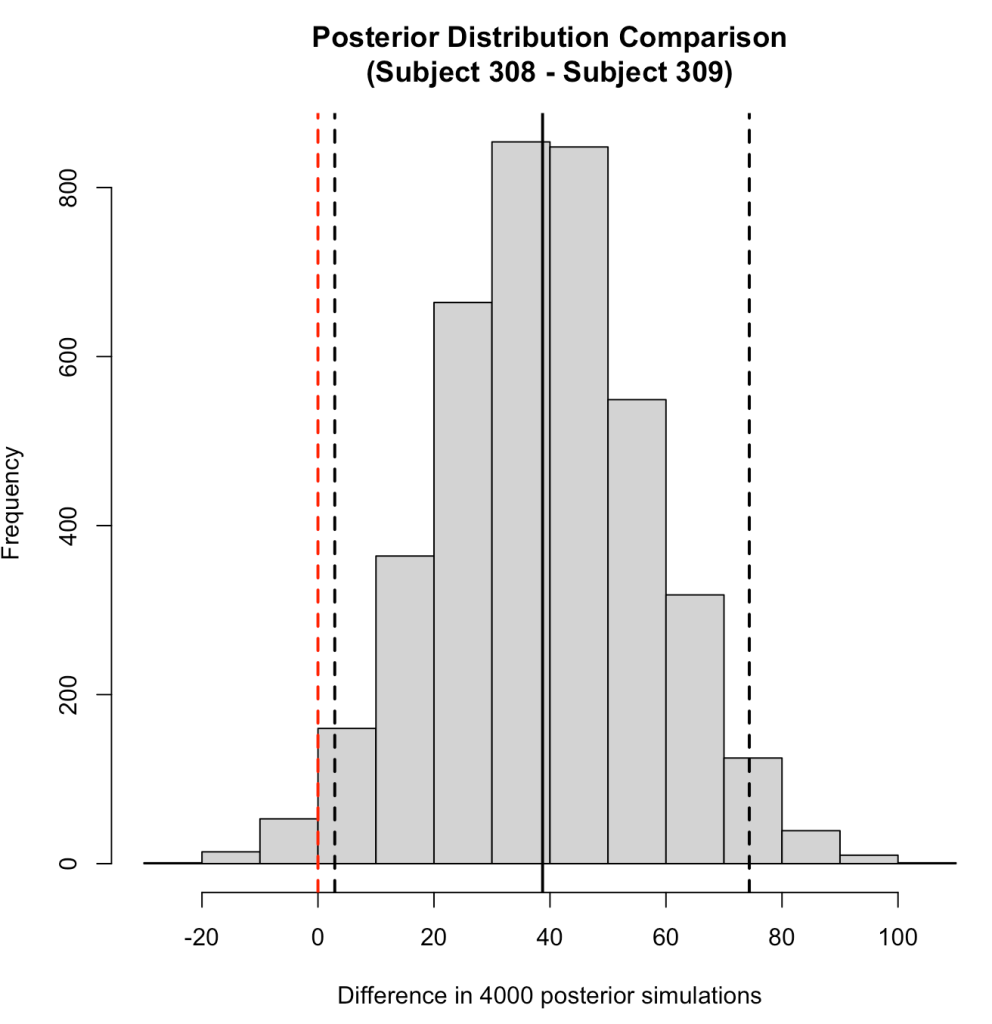
We can also use the posterior distributions to make a probabilistic statement about how often, in the 4000 posterior draws, Subject 308 had a higher reaction time than Subject 309.
prop.table(table(posterior_intercept_sims[, 1] > posterior_intercept_sims[, 2]))
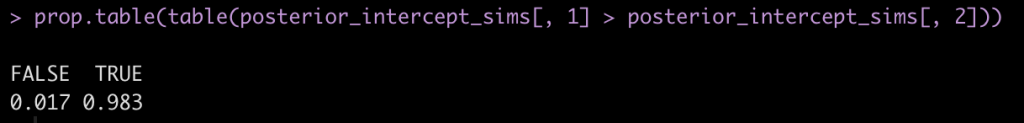
Here we see that mean posterior probability that Subject 308 has a higher reaction time than Subject 309 is 98.3%.
Of course we could (and should) go through and do a similar work up for the slope coefficient for the Days variable. However, this is getting long, so I’ll leave you to try that out on your own.
Wrapping Up
Mixed models are an interesting way of handling data consisting of multiple measurements taken on different individuals, as common in sport science. Thanks to Newans et al (1) for sharing their insights into these models. Hopefully this blog was useful in explaining a little bit about how mixed models work and how extending them to a Bayesian framework offers some additional ways of looking at the data. Just note that if you run the code on your own, the Bayesian results might be slightly different than mine as the Monte Carlo component of the Bayesian model produces some randomness in each simulation (and I didn’t set a seed prior to running the model).
The entire code is available on my GitHub page.
If you notice any errors in the code, please reach out!
References
1. Newans T, Bellinger P, Drovandi C, Buxton S, Minahan C. (2022). The utility of mixed models in sport science: A call for further adoption in longitudinal data sets. Int J Sports Phys Perf. Published ahead of print.
2. Matthews JNS, Altman DG, Campbell MJ, Royston P. (1990). Analysis of serial measurements in medical research. Br Med J; 300: 230-235.
3. Weston M, Drust B, Gregson W. (2011). Intensities of exercise during match-play in FA Premier League Referees and players. J Sports Sc; 29(5): 527-532.
4. Gelman A, Hill J. (2009). Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge University Press.